Stat 470/670 Lecture 25: Multi-Dimensional
Scaling
Julia Fukuyama
Setup for multi-dimensional scaling
Instead of measurements on variables, like in PCA, we have distances
between the samples.
The distances can be what was measured initially, or the distance
could be constructed by the analyst from other variables that were
measured directly.
In multi-dimensional scaling, the goal is to make a map of the
samples in a low-dimensional space (probably 2-dimensional space) so
that the distances in that map match the distances between the samples
as closely as possible.
Fake Data 1
D = matrix(c(0, 1, sqrt(2),
1, 0, 1,
sqrt(2), 1, 0),
nrow = 3, byrow = TRUE)
round(D, digits = 2)
## [,1] [,2] [,3]
## [1,] 0.00 1 1.41
## [2,] 1.00 0 1.00
## [3,] 1.41 1 0.00
Here D is a distance matrix, and the \((i,j)\) element of D tells us
the distance between sample \(i\) and
sample \(j\).
How would you position these samples in space so that the distances
between them matched the distances in D?
Embedding into Euclidean space
In multi-dimensional scaling, we want to find an embedding
of the samples into Euclidean space so that the distances between the
embedded points match the distances between the samples as closely as
possible.
This sounds fancy, but all it means is that we create a set of
coordinates and assign each sample a value along each coordinate so that
the distances between the samples match the input distances.
How does this work on our fake data?
## [,1] [,2]
## [1,] 7.071068e-01 0.2357023
## [2,] -1.110223e-16 -0.4714045
## [3,] -7.071068e-01 0.2357023
mds_points = cmdscale(D)
ggplot(data.frame(mds_points)) +
geom_text(aes(x = X1, y = X2, label = 1:3)) + coord_fixed()
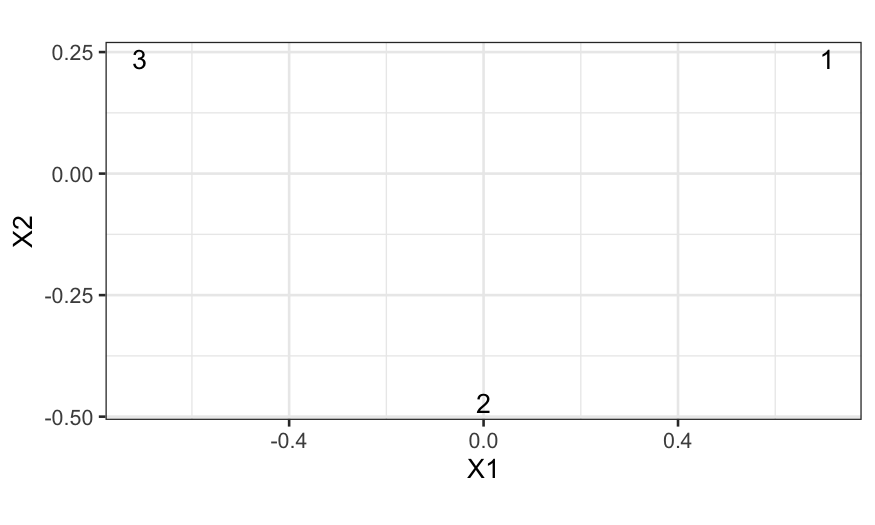
We can check that the distances match:
## The dist function computes distances (Euclidean
## by default) between the rows of a data frame
dist(mds_points)
## 1 2
## 2 1.000000
## 3 1.414214 1.000000
## compare with D, the input distances
D
## [,1] [,2] [,3]
## [1,] 0.000000 1 1.414214
## [2,] 1.000000 0 1.000000
## [3,] 1.414214 1 0.000000
Classical Multi-Dimensional Scaling
One solution to the multi-dimensional scaling problem is given by
classical multi-dimensional scaling.
Let \(\mathbf D \in \mathbb R^{n \times
n}\) be a matrix where \(\mathbf
D_{ij}\) contains the square of the distance between sample \(i\) and sample \(j\).
Let \(\mathbf H \in \mathbb R^{n \times
n}\) be the centering matrix, \(\mathbf
H = \mathbf I - \frac{1}{n} \mathbf 1 \mathbf 1^T\).
Create the doubly-centered distance matrix \(\mathbf B = -\frac{1}{2} \mathbf H \mathbf D
\mathbf H\), and let \(\mathbf U
\mathbf \Lambda \mathbf U^T\) be the singular value decomposition
of \(\mathbf B\).
Then the \(k\)-dimensional solution
to the multi-dimensional scaling problem is obtained by taking \(\mathbf U_{(k)} \mathbf
\Lambda_{(k)}^{1/2}\).
Idea behind this solution:
Suppose the distance really did come from a matrix \(\mathbf X \in \mathbb R^{n \times k}\),
where we computed the Euclidean distances between the rows of \(\mathbf X\). For definiteness, assume that
the columns of \(\mathbf X\) are
centered.
Then it turns out (linear algebra exercise: verify this) that \((\mathbf H \mathbf X) (\mathbf H \mathbf X)^T =
\mathbf B\).
The top \(k\) left singular vectors
of \(\mathbf X\) (which is the same as
\(\mathbf H \mathbf X\) because \(\mathbf X\) already has centered columns)
will therefore give the optimal representation of the true embedded
points that we got the distances from.
The singular vectors of \(\mathbf H \mathbf
X\) are the same as the singular vectors of \(\mathbf B\), so if we start off with \(\mathbf B\) instead of \(\mathbf X\), we can still get the optimal
low-dimensional embedding by taking the top singular vectors of \(\mathbf B\).
Checking the quality of the MDS solution
Just as in PCA and with the SVD, we have a measure of the quality of
the approximation.
In classical multi-dimensional scaling, these are given by the
eigenvalues of \(\mathbf B\), and
plotting the eigenvalues tells us how how much of the “variance” is
explained by the multi-dimensional scaling axes.
If we can represent the distances perfectly with an embedding into
\(k\)-dimensional space, the top \(k\) eigenvalues will be non-zero and the
remainder will be zero.
We can check this on our fake data, where we constructed the
distances so that they could be exactly represented in two-dimensional
space.
cmdscale(D, eig = TRUE)$eig
## [1] 1.000000e+00 3.333333e-01 2.220446e-16
In general, you won’t be able to get an exact representation in a
number of dimensions that’s easy to visualize, but you will want to know
how well you’re doing with the number of dimensions you take.
We use the eigenvalues to make a scree plot, analogous to the PCA
scree plot, to measure the quality of the embedding
Major difference between MDS and PCA:.
- The eigenvalues can be negative.
- Negative eigenvalues mean that there is no embedding of the points
so that the Euclidean distances between them exactly match the input
distances, and the size of the negative eigenvalues indicate how severe
the problem is.
- Not that important, but the terminology is that if you see negative
eigenvalues, it means that your distances are
non-Euclidean.
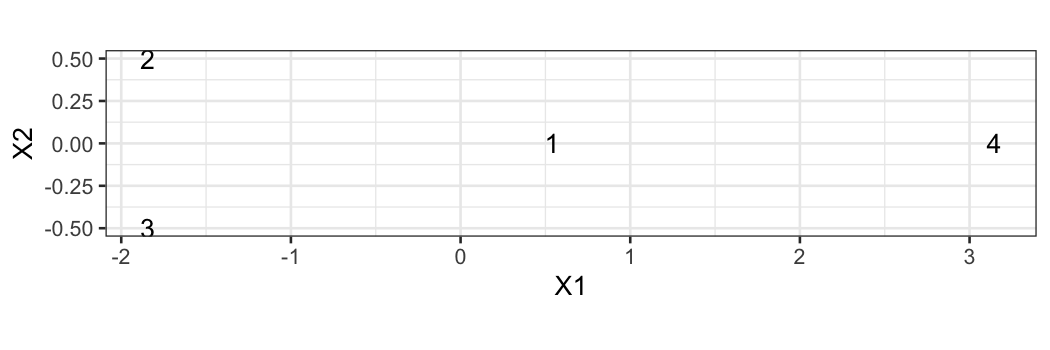
Example of non-embeddable set of points
D = matrix(c(0, 1, 1, .1,
1, 0, 1, 5,
1, 1, 0, 5,
.1, 5, 5, 0),
nrow = 4, byrow = TRUE)
D
## [,1] [,2] [,3] [,4]
## [1,] 0.0 1 1 0.1
## [2,] 1.0 0 1 5.0
## [3,] 1.0 1 0 5.0
## [4,] 0.1 5 5 0.0
mds_points = data.frame(cmdscale(D))
dist(mds_points)
## 1 2 3
## 2 2.436166
## 3 2.436166 1.000000
## 4 2.605269 5.014562 5.014562
ggplot(mds_points) +
geom_text(aes(x = X1, y = X2, label = 1:4)) + coord_fixed()
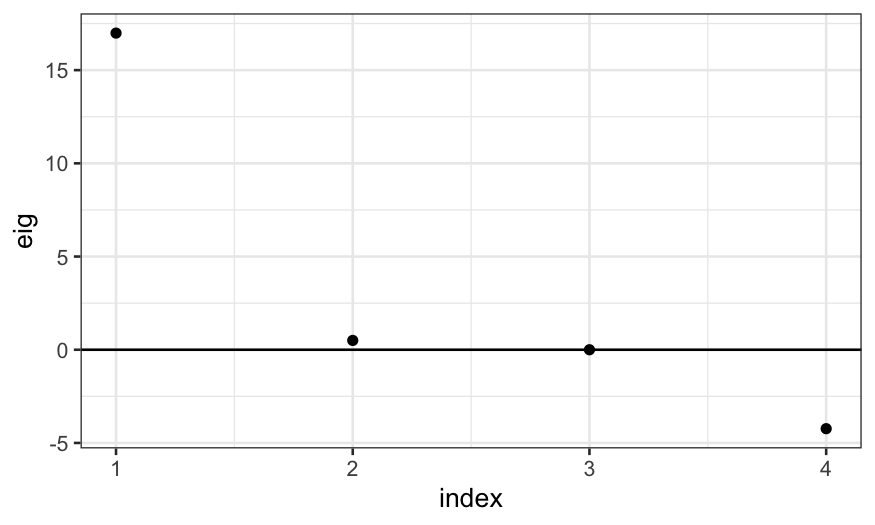
As promised, this shows up in negative eigenvalues:
mds_eig = data.frame(eig = cmdscale(D, eig = TRUE)$eig, index = 1:4)
mds_eig
## eig index
## 1 16.987227 1
## 2 0.500000 2
## 3 0.000000 3
## 4 -4.234727 4
ggplot(mds_eig) + geom_point(aes(x = index, y = eig)) + geom_hline(yintercept = 0)
Contrived Example 1: State locations
R contains data on state locations, including one called
state.center that gives the latitude and longitude of the
center of each state.
What happens if we compute distances between the centers of the
states and run multi-dimensional scaling on those distances?
state_locations = data.frame(state.center)
state_distances = dist(state_locations, method = "euclidean")
state_mds = cmdscale(state_distances, eig = TRUE, k = 2)
Before we get to the MDS plot, let’s look at the scree plot to see
the quality of the MDS solution.
ggplot(data.frame(eig = state_mds$eig, index = 1:50)) +
geom_point(aes(x = index, y = eig)) +
ggtitle("Scree plot for MDS on distances between states")
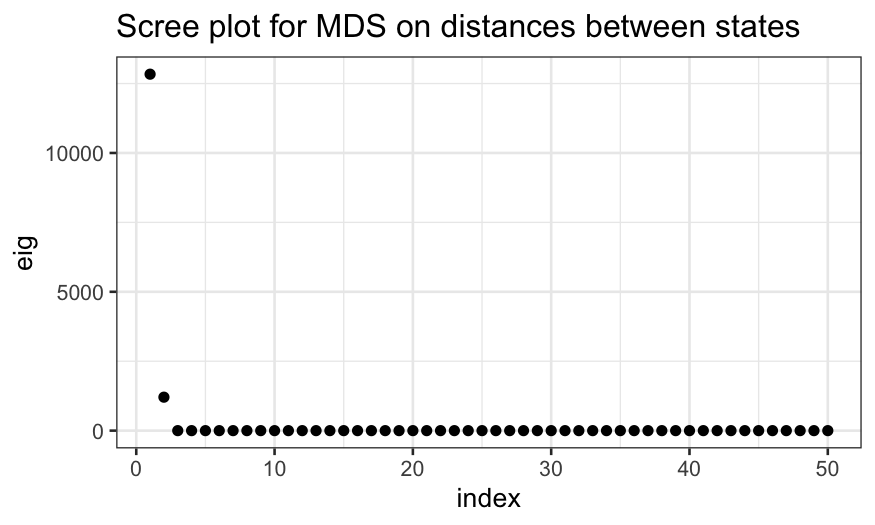
Why do we only get non-zero eigenvalues for the first two MDS
axes?
ggplot(data.frame(state_mds$points), aes(x = X1, y = X2, label = state.name)) +
geom_text_repel() + geom_point() + coord_fixed()
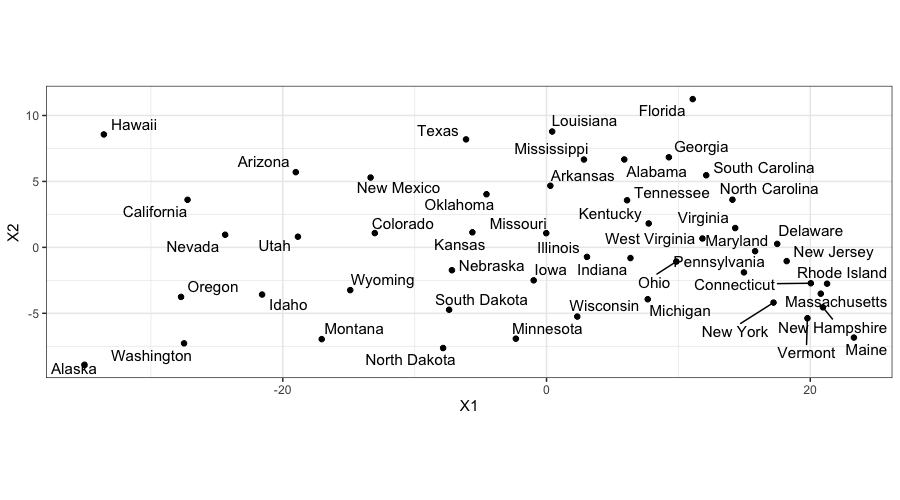
When we plot the MDS solution, we get a map!
The states all have the correct relative locations, but the
north-south axis is going the wrong way.
This is just due to an indeterminacy in the solution: the singular
value decomposition is only determined up to a sign change for the
singular vectors.
More heuristically, since we only provide MDS with distances, we can
only expect it to give us good approximations to the distances between
the samples, we can’t expect it to know about north and south.
Non-metric MDS
Non-metric MDS is a robust alternative to classical MDS, and it is
used when we want a map that preserves relative distances instead of
absolute distances.
The idea is that we want to find a an embedding of the points into a
lower-dimensional space so that the ranks of the distances are preserved
as well as possible (the points that are the farthest from each other in
the embedded space have the largest input distance, the points that are
closest to each other in the embedded space have the smallest input
distance, etc.).
To do this, we find an embedding of the points into a
lower-dimensional space and a monotonic transformation of the
embedded distances so that the transformed distances recapitulate the
input distances as well as possible. The monotone transformation is
essentially a trick that allows us to match ranks of distances instead
of absolute distances.
Notes:
- NMDS is more resistant to outliers than classical MDS: if one point
has a very large distance from all the others, the first classical MDS
axis will tend to separate that point from the others and not be
informative about the remaining distances.
- Unlike classical MDS, NMDS does not give nested solutions: if we do
NMDS with 2 axes, the first axis will not be equal to the NMDS solution
with 1 axis.
- There is no notion of percentage of variation explained by
individual axes as in classical MDS.
Implementation of NMDS
Let \(d_i\) contain the input
distances, and let \(f\) be a monotone
increasing function.
Note that since \(f\) is monotone,
\(d_i < d_j\) implies that \(f(d_i) < f(d_j)\), and so the
relative distances between the points are preserved under \(f\).
In NMDS, we want to minimize the stress function, defined as \[
\text{STRESS}^2 = \frac{\sum_i (f(\tilde d_i) - d_i)^2}{\sum_j d_j^2}
\] where \(d\) represents the
input distances, and \(\tilde d\)
represent the distances between the embedded points.
The NMDS algorithm is as follows:
- Find a random embedding of the samples, e. g. by sampling from a
normal distribution.
- Calculate the distances \(\tilde
d\) between the embedded sample points.
- Find the optimal monotonic transformation of the distances \(f\) so that \(f(\tilde d)\) matches \(d\) as closely as possible.
- Find the embedding of the samples such that the distances between
the embedded points matches \(f(d)\) as
closely as possible.
- Compare the stress to some criterion. If the change in stress is
small enough then exit the algorithm, otherwise return to to step
2.
Example on colors
In a psychology study (Ekman, Gosta. 1954. “Dimensions of Color
Vision.” The Journal of Psychology 38 (2). Taylor & Francis:
467–74.), the investigator asked subjects to rate similarities between
colors.
These were combined to give overall measure of similarities between
colors, and the results are in ekman.txt.
ekm = read.table("../../datasets/ekman.txt", header=TRUE)
rownames(ekm) = colnames(ekm)
## the elements in ekm are similarities, but we
## need dissimilarities instead. We can create
## dissimilarities by taking the complement of
## the similarities and setting the diagonal to zero.
ekm_dist = 1 - ekm - diag(1, ncol(ekm))
ekm_dist[1:5, 1:5]
## w434 w445 w465 w472 w490
## w434 0.00 0.14 0.58 0.58 0.82
## w445 0.14 0.00 0.50 0.56 0.78
## w465 0.58 0.50 0.00 0.19 0.53
## w472 0.58 0.56 0.19 0.00 0.46
## w490 0.82 0.78 0.53 0.46 0.00
Let’s try classical MDS first:
ekm_mds = cmdscale(ekm_dist, eig = TRUE)
## we can make a scree plot giving
## the fraction of variance explained
ggplot(data.frame(eig = ekm_mds$eig,
index = 1:length(ekm_mds$eig))) +
geom_point(aes(x = index, y = eig))
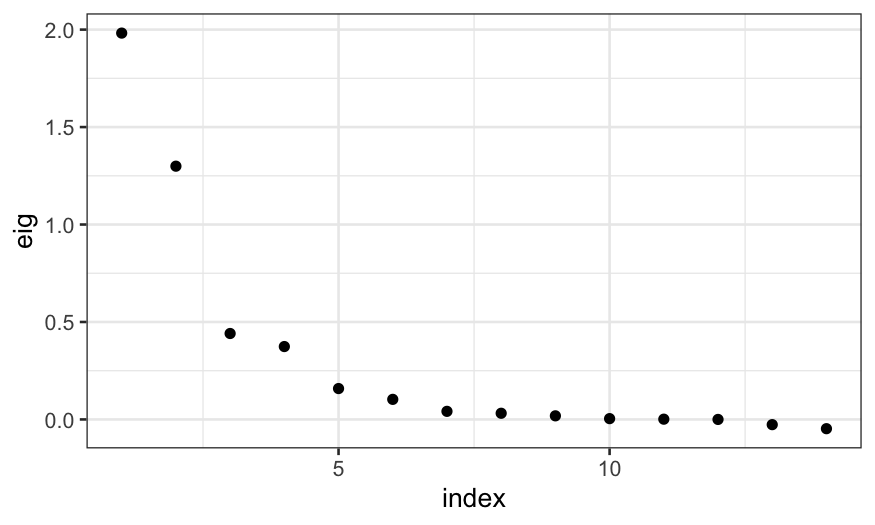
Note that the negative eigenvalues at the end indicate that the
dissimilarities cannot be exactly represented in Euclidean space, but
the values are not that large and so we aren’t too concerned.
The top two eigenvalues are quite large, indicating that a
two-dimensional MDS solution does a reasonable job at recapitulating the
dissimilarities between the samples.
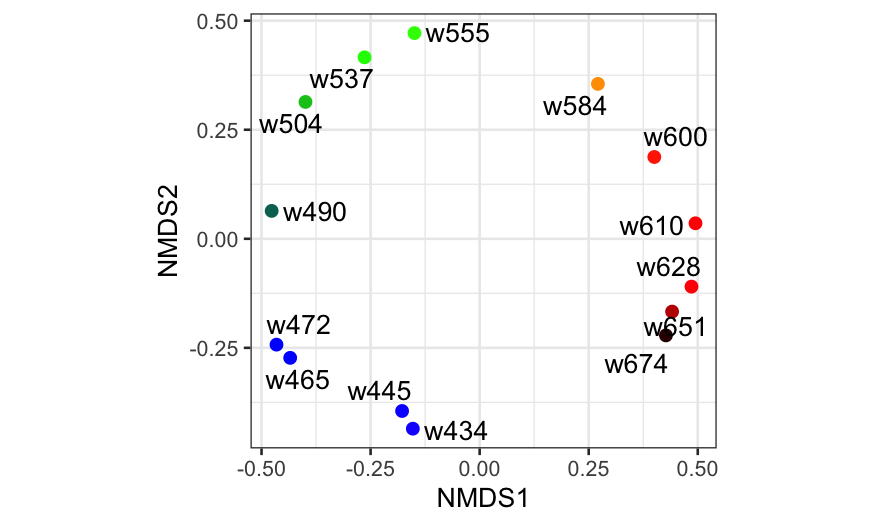
And finally the plot:
## Here we're changing variable names and
## adding some additional information to
## the data frame we will use to plot the
## MDS solution
ekm_points = ekm_mds$points[,1:2] %>%
as_tibble %>%
setNames(paste0("MDS", 1:2)) %>%
mutate(
name = rownames(ekm),
rgb = photobiology::w_length2rgb(as.numeric(sub("w", "", name))))
ggplot(ekm_points, aes(x = MDS1, y = MDS2)) +
geom_point(col = ekm_points$rgb, size = 2) +
geom_text_repel(aes(label = name)) + coord_fixed()
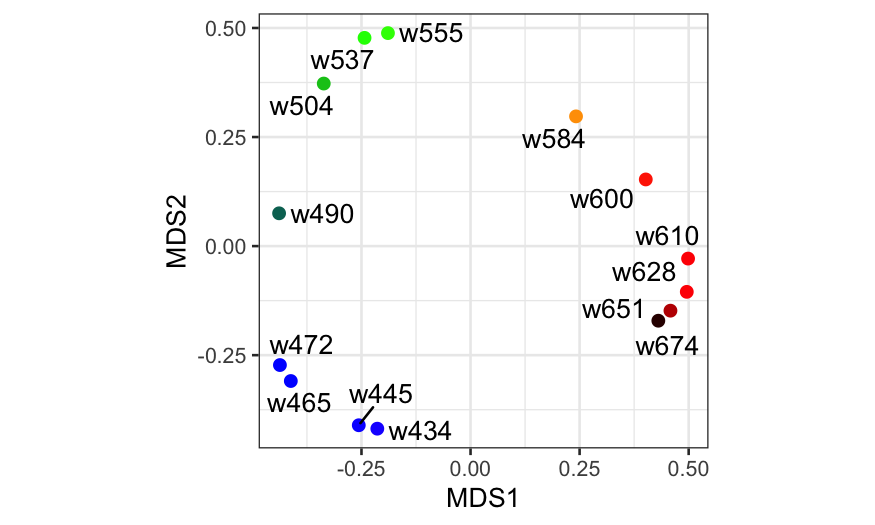
Color data with NMDS
Remember that NMDS is a randomized algorithm, so each run can in
principle give a different solution.
The metaMDS function (in the package vegan)
repeats the NMDS algorithm many times (20 by default) and looks for the
best solution among the results.
The output here tells us that for each run of the algorithm, the
stresses are about the same, suggesting that the corresponding solutions
are the same.
The part of the output that says
Procrustes: rmse 1.060348e-06 max resid
1.960065e-06
is checking for similarity between the solutions directly:
rmse and max resid describe the discrepancies
between the solution and the previous best solution.
Here we see that the algorithm converges ot the same solution every
time.
ekm_nmds = metaMDS(ekm_dist, k = 2, autotransform = FALSE)
## Run 0 stress 0.02310251
## Run 1 stress 0.02310251
## ... Procrustes: rmse 2.102867e-06 max resid 3.320966e-06
## ... Similar to previous best
## Run 2 stress 0.02310251
## ... Procrustes: rmse 7.088863e-06 max resid 1.145484e-05
## ... Similar to previous best
## Run 3 stress 0.02310251
## ... Procrustes: rmse 6.438299e-06 max resid 1.040306e-05
## ... Similar to previous best
## Run 4 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 4.681247e-06 max resid 7.620905e-06
## ... Similar to previous best
## Run 5 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 3.633134e-06 max resid 6.162556e-06
## ... Similar to previous best
## Run 6 stress 0.02310251
## ... Procrustes: rmse 3.021893e-06 max resid 5.156862e-06
## ... Similar to previous best
## Run 7 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 1.146055e-06 max resid 2.470557e-06
## ... Similar to previous best
## Run 8 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 9.659437e-07 max resid 2.036998e-06
## ... Similar to previous best
## Run 9 stress 0.02310251
## ... Procrustes: rmse 4.787886e-06 max resid 8.130221e-06
## ... Similar to previous best
## Run 10 stress 0.02310251
## ... Procrustes: rmse 5.616317e-07 max resid 8.747772e-07
## ... Similar to previous best
## Run 11 stress 0.02310251
## ... Procrustes: rmse 5.527651e-07 max resid 1.104713e-06
## ... Similar to previous best
## Run 12 stress 0.02310251
## ... Procrustes: rmse 2.855771e-06 max resid 4.601952e-06
## ... Similar to previous best
## Run 13 stress 0.02310251
## ... Procrustes: rmse 1.506129e-06 max resid 2.440185e-06
## ... Similar to previous best
## Run 14 stress 0.02310251
## ... Procrustes: rmse 6.926222e-06 max resid 1.15127e-05
## ... Similar to previous best
## Run 15 stress 0.02310251
## ... Procrustes: rmse 8.158106e-07 max resid 1.29539e-06
## ... Similar to previous best
## Run 16 stress 0.02310251
## ... Procrustes: rmse 1.780811e-06 max resid 2.655272e-06
## ... Similar to previous best
## Run 17 stress 0.02310251
## ... Procrustes: rmse 6.859623e-07 max resid 1.288504e-06
## ... Similar to previous best
## Run 18 stress 0.02310251
## ... Procrustes: rmse 5.879151e-07 max resid 8.94391e-07
## ... Similar to previous best
## Run 19 stress 0.02310251
## ... Procrustes: rmse 4.007526e-06 max resid 6.695112e-06
## ... Similar to previous best
## Run 20 stress 0.02310251
## ... Procrustes: rmse 3.797161e-06 max resid 5.754955e-06
## ... Similar to previous best
## *** Best solution repeated 13 times
We would like an analog of the scree plot so that we can evaluate how
many dimensions to use.
One way to do this is to compute the stress function for each number
of dimensions and plot that.
## since the algorithm is random, it would be
## better to do this many times for each value
## of k and take the average
stresses = sapply(1:5, function(k) metaMDS(ekm_dist, k = k, autotransform = FALSE)$stress)
## Run 0 stress 0.2721258
## Run 1 stress 0.5212407
## Run 2 stress 0.4856468
## Run 3 stress 0.490962
## Run 4 stress 0.2567348
## ... New best solution
## ... Procrustes: rmse 0.05564529 max resid 0.1384309
## Run 5 stress 0.2687508
## Run 6 stress 0.4533083
## Run 7 stress 0.4373748
## Run 8 stress 0.4843117
## Run 9 stress 0.5071186
## Run 10 stress 0.5291604
## Run 11 stress 0.4866729
## Run 12 stress 0.4983676
## Run 13 stress 0.4942408
## Run 14 stress 0.4915839
## Run 15 stress 0.4584145
## Run 16 stress 0.5006447
## Run 17 stress 0.5165007
## Run 18 stress 0.5246208
## Run 19 stress 0.5003296
## Run 20 stress 0.5081357
## *** Best solution was not repeated -- monoMDS stopping criteria:
## 20: scale factor of the gradient < sfgrmin
## Run 0 stress 0.02310251
## Run 1 stress 0.02310251
## ... Procrustes: rmse 1.603649e-06 max resid 2.608348e-06
## ... Similar to previous best
## Run 2 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 1.283596e-06 max resid 2.203181e-06
## ... Similar to previous best
## Run 3 stress 0.02310251
## ... Procrustes: rmse 9.753636e-07 max resid 1.832654e-06
## ... Similar to previous best
## Run 4 stress 0.2925016
## Run 5 stress 0.02310251
## ... Procrustes: rmse 5.337933e-06 max resid 8.633325e-06
## ... Similar to previous best
## Run 6 stress 0.02310251
## ... Procrustes: rmse 1.579965e-06 max resid 3.45673e-06
## ... Similar to previous best
## Run 7 stress 0.02310251
## ... Procrustes: rmse 3.284343e-06 max resid 5.161338e-06
## ... Similar to previous best
## Run 8 stress 0.02310251
## ... Procrustes: rmse 2.555243e-06 max resid 4.933159e-06
## ... Similar to previous best
## Run 9 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 1.01295e-06 max resid 2.103083e-06
## ... Similar to previous best
## Run 10 stress 0.02310251
## ... Procrustes: rmse 2.232036e-06 max resid 4.119736e-06
## ... Similar to previous best
## Run 11 stress 0.02310251
## ... Procrustes: rmse 1.663214e-06 max resid 3.072049e-06
## ... Similar to previous best
## Run 12 stress 0.02310251
## ... Procrustes: rmse 2.237823e-06 max resid 4.174616e-06
## ... Similar to previous best
## Run 13 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 1.327781e-06 max resid 2.668128e-06
## ... Similar to previous best
## Run 14 stress 0.02310251
## ... Procrustes: rmse 3.930095e-06 max resid 6.673939e-06
## ... Similar to previous best
## Run 15 stress 0.02310251
## ... Procrustes: rmse 1.227083e-06 max resid 2.181569e-06
## ... Similar to previous best
## Run 16 stress 0.02310251
## ... Procrustes: rmse 1.61893e-06 max resid 2.611836e-06
## ... Similar to previous best
## Run 17 stress 0.02310251
## ... Procrustes: rmse 4.812507e-06 max resid 7.680302e-06
## ... Similar to previous best
## Run 18 stress 0.02310251
## ... New best solution
## ... Procrustes: rmse 9.04642e-07 max resid 1.366657e-06
## ... Similar to previous best
## Run 19 stress 0.02310251
## ... Procrustes: rmse 2.685811e-06 max resid 5.192665e-06
## ... Similar to previous best
## Run 20 stress 0.02310251
## ... Procrustes: rmse 4.933418e-06 max resid 7.938065e-06
## ... Similar to previous best
## *** Best solution repeated 3 times
## Run 0 stress 0.01253494
## Run 1 stress 0.01703834
## Run 2 stress 0.1085453
## Run 3 stress 0.01715047
## Run 4 stress 0.01483193
## Run 5 stress 0.0156638
## Run 6 stress 0.01253473
## ... New best solution
## ... Procrustes: rmse 0.000744619 max resid 0.00139024
## ... Similar to previous best
## Run 7 stress 0.01253767
## ... Procrustes: rmse 0.0008841715 max resid 0.001456
## ... Similar to previous best
## Run 8 stress 0.01678396
## Run 9 stress 0.0171508
## Run 10 stress 0.01529306
## Run 11 stress 0.01529293
## Run 12 stress 0.01253648
## ... Procrustes: rmse 0.001276111 max resid 0.002392105
## ... Similar to previous best
## Run 13 stress 0.01619717
## Run 14 stress 0.01514985
## Run 15 stress 0.01617106
## Run 16 stress 0.012539
## ... Procrustes: rmse 0.001555727 max resid 0.00261888
## ... Similar to previous best
## Run 17 stress 0.0160428
## Run 18 stress 0.01777524
## Run 19 stress 0.0140412
## Run 20 stress 0.0124441
## ... New best solution
## ... Procrustes: rmse 0.01877198 max resid 0.04687448
## *** Best solution was not repeated -- monoMDS stopping criteria:
## 11: no. of iterations >= maxit
## 9: stress ratio > sratmax
## Run 0 stress 0.006273768
## Run 1 stress 0.004990382
## ... New best solution
## ... Procrustes: rmse 0.1007591 max resid 0.1849666
## Run 2 stress 0.00487154
## ... New best solution
## ... Procrustes: rmse 0.01068611 max resid 0.02011363
## Run 3 stress 0.006313356
## Run 4 stress 0.005815049
## Run 5 stress 0.005724101
## Run 6 stress 0.01109444
## Run 7 stress 0.003030431
## ... New best solution
## ... Procrustes: rmse 0.04301309 max resid 0.07967849
## Run 8 stress 0.003766835
## Run 9 stress 0.002812255
## ... New best solution
## ... Procrustes: rmse 0.007655836 max resid 0.01192655
## Run 10 stress 0.002872299
## ... Procrustes: rmse 0.01055606 max resid 0.01956895
## Run 11 stress 0.006120003
## Run 12 stress 0.1316872
## Run 13 stress 0.003452073
## Run 14 stress 0.005912479
## Run 15 stress 0.006849582
## Run 16 stress 0.003751898
## Run 17 stress 0.006483368
## Run 18 stress 0.003709185
## Run 19 stress 0.004488317
## Run 20 stress 0.002733593
## ... New best solution
## ... Procrustes: rmse 0.008442853 max resid 0.01632713
## *** Best solution was not repeated -- monoMDS stopping criteria:
## 19: no. of iterations >= maxit
## 1: stress ratio > sratmax
## Run 0 stress 0.0009367835
## Run 1 stress 0.002131138
## Run 2 stress 0.003907969
## Run 3 stress 0.001706481
## Run 4 stress 0.005466837
## Run 5 stress 0.002919591
## Run 6 stress 0.00258284
## Run 7 stress 0.003337378
## Run 8 stress 0.005895784
## Run 9 stress 0.001076621
## ... Procrustes: rmse 0.09829673 max resid 0.2129623
## Run 10 stress 0.002311725
## Run 11 stress 0.002533747
## Run 12 stress 0.001741355
## Run 13 stress 0.002134076
## Run 14 stress 0.001557201
## Run 15 stress 0.0006962816
## ... New best solution
## ... Procrustes: rmse 0.08041192 max resid 0.1217456
## Run 16 stress 0.001892428
## Run 17 stress 0.002917626
## Run 18 stress 0.001702046
## Run 19 stress 0.002460906
## Run 20 stress 0.002818138
## *** Best solution was not repeated -- monoMDS stopping criteria:
## 20: no. of iterations >= maxit
## note about above, autotransform is a parameter
## that is specific to ecology data, we want to
## set it to FALSE
ggplot(data.frame(stresses, k = 1:5)) + geom_point(aes(x = k, y = stresses))
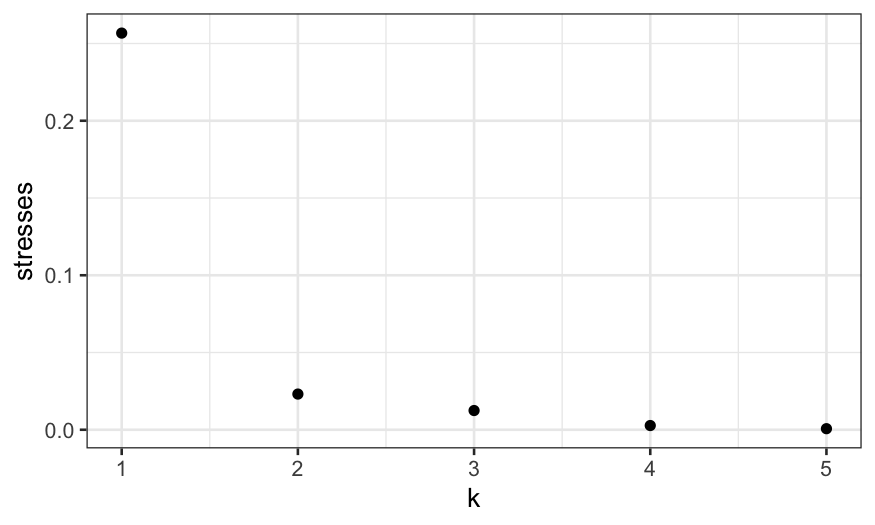
Here we see that the stress drops substantially going from 1 to 2
dimensions, and then doesn’t go down that much once we increase the
number of dimensions further.
This indicates to us that the two-dimensional solution is likely to
be pretty good, just as it was with classical MDS.
And the plot:
nmds_points = ekm_nmds$points[,1:2] %>%
as_tibble %>%
setNames(paste0("NMDS", 1:2)) %>%
bind_cols(select(ekm_points, rgb, name))
ggplot(nmds_points, aes(x = NMDS1, y = NMDS2)) +
geom_point(col = ekm_points$rgb, size = 2) +
geom_text_repel(aes(label = name)) + coord_fixed()