Stat 470/670 Lecture 24: Scree plots and choosing
the number of dimensions
Julia Fukuyama
Today
Scree plot and how we interpret it
What does the scree plot look like when the data matrix is just
noise
How do we choose a good number of dimensions for PCA
Scree plots
In a scree plot, we plot the eigenvalues in decreasing
order.
It is used to help us choose the number of “important” principal
components.
Scree plot: Advice
- Look for an elbow (the plot is supposed to look like a scree).
- Take as many components as necessary to get to at least some
pre-determined fraction of variance explained.
- Compare eigenvalues to a null distribution.
Three simulated examples
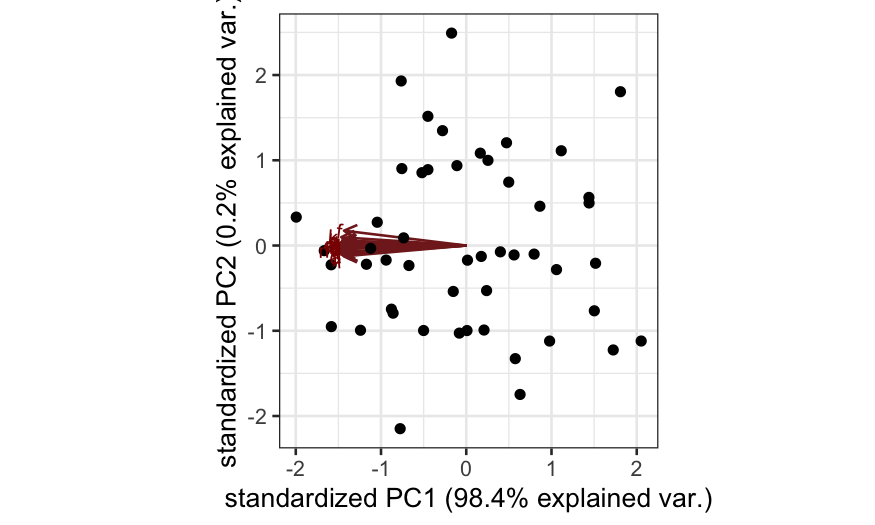
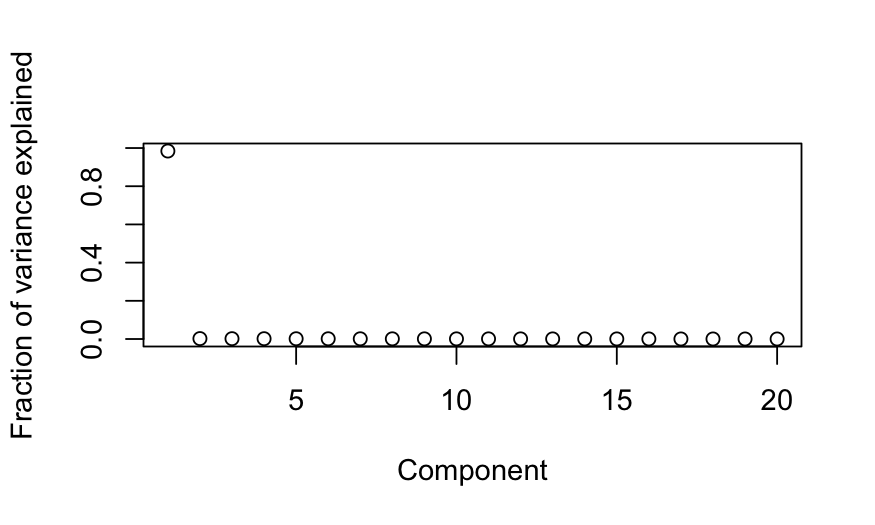
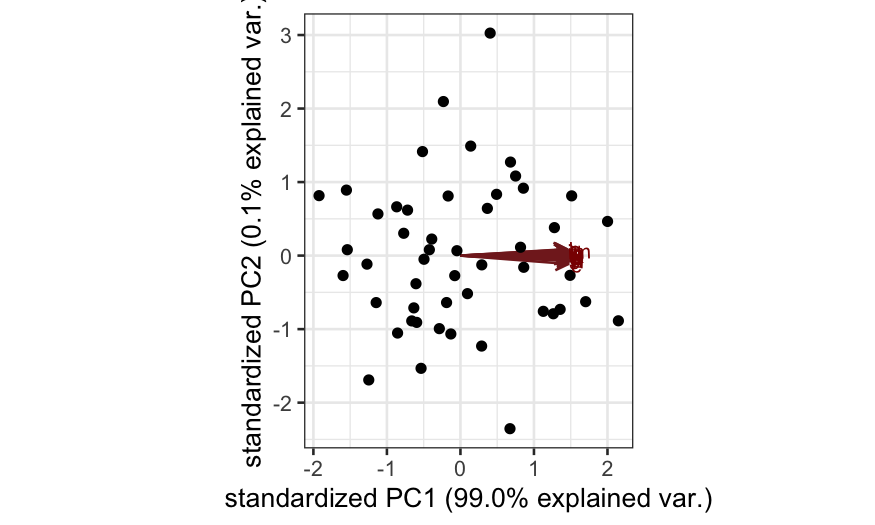
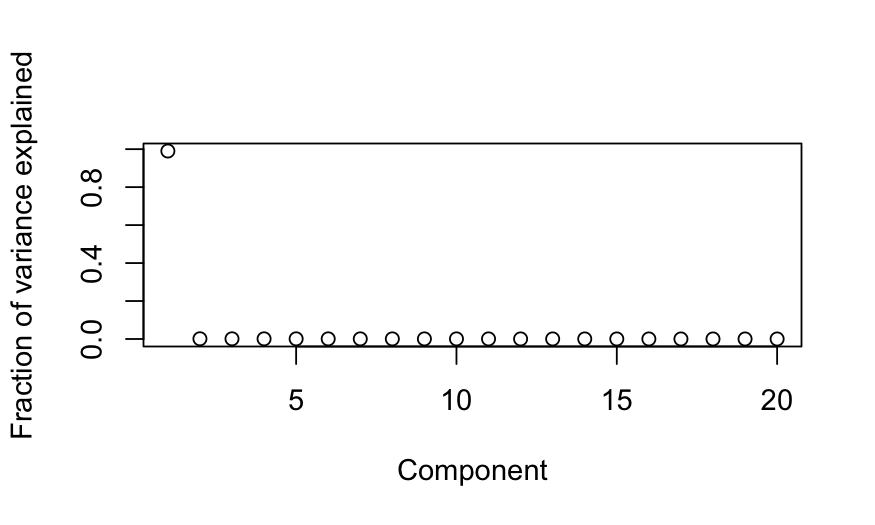
- Simulation A: All of the variables are highly correlated with each
other. Interpretation: all of the variables are measuring the same
underlying quantity.
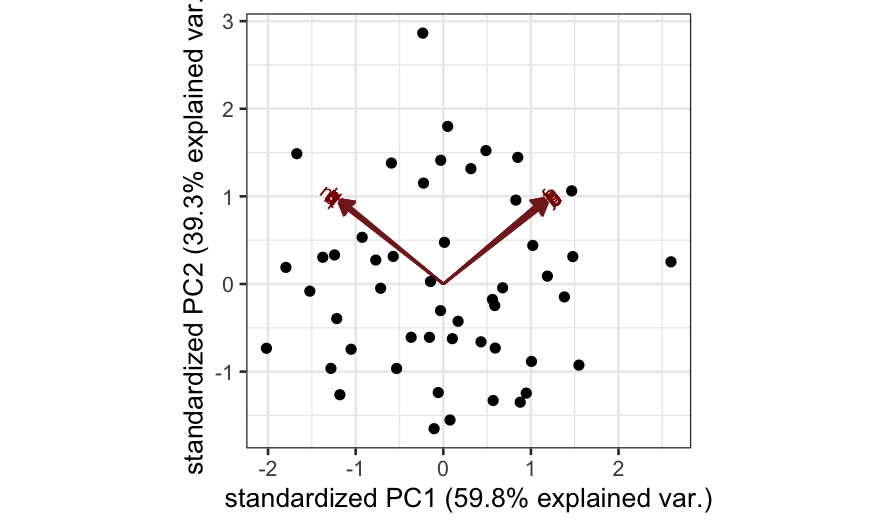
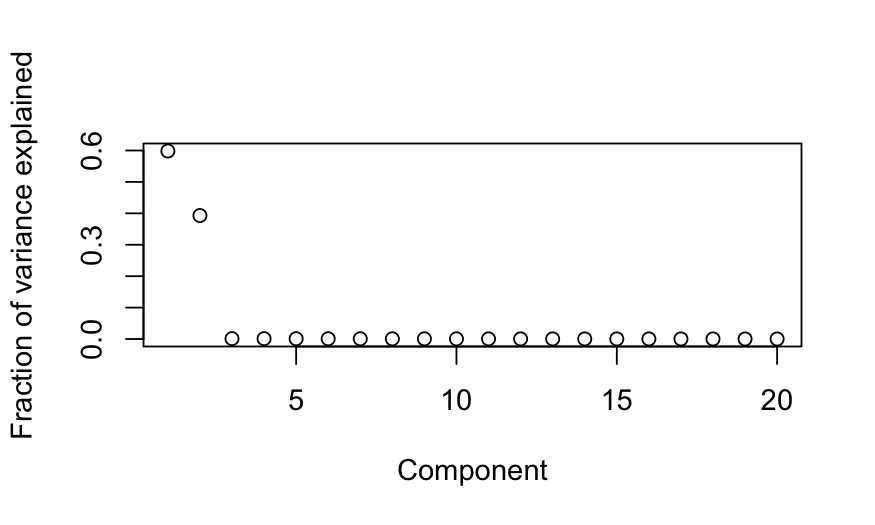
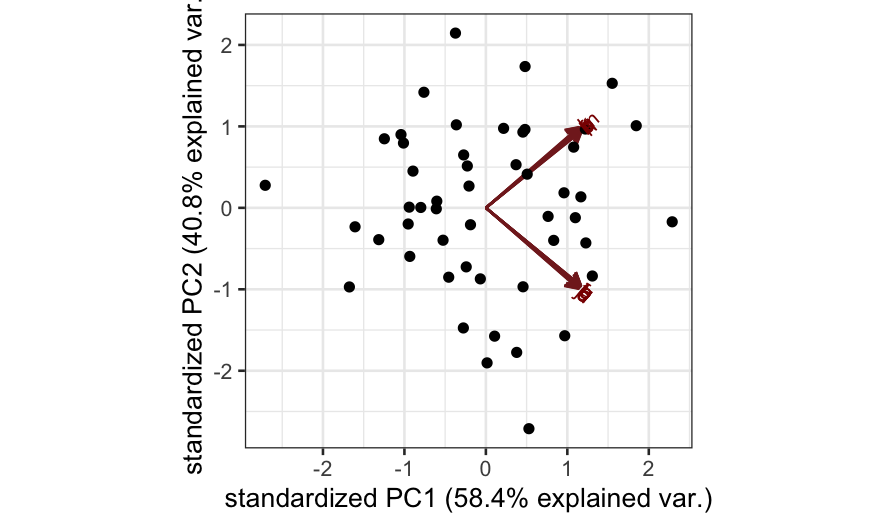
- Simulation B: There are two blocks of variables, with high
correlations within each block. Interpretation: there are two
independent underlying factors, and each of the variables measures one
of them.
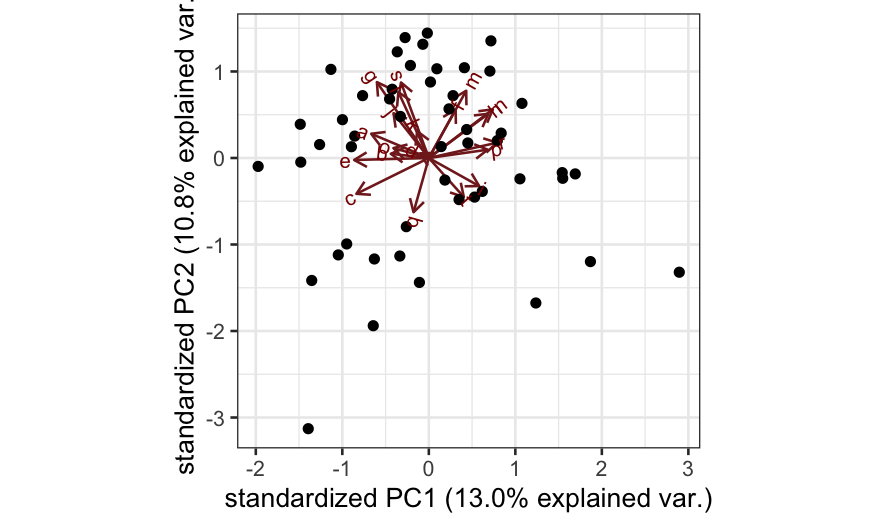
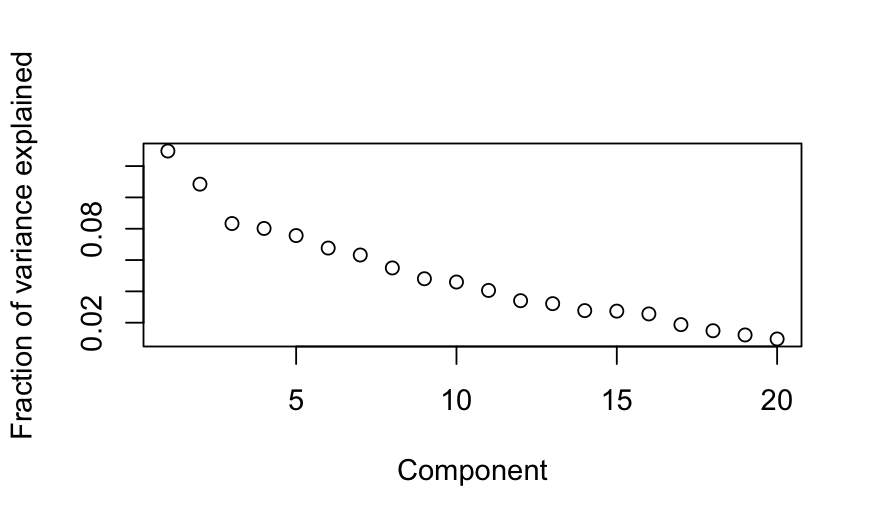
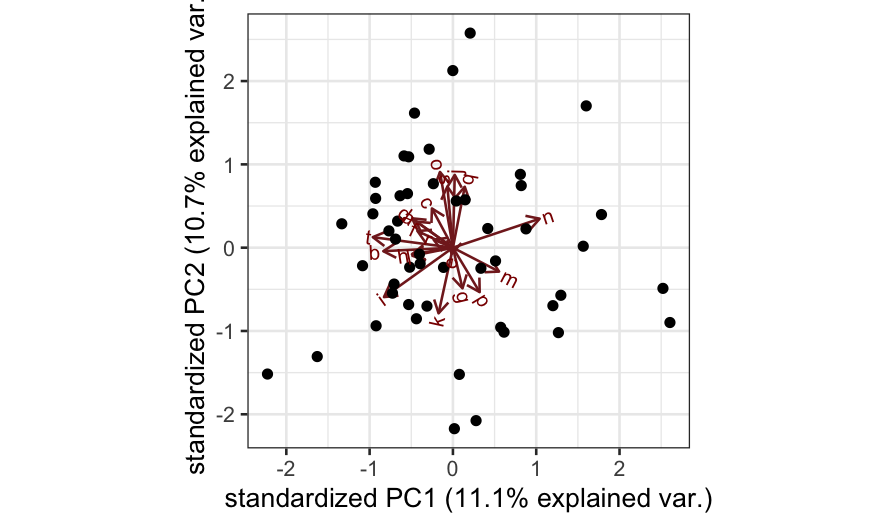
- Simulation C: All of the variables are independent. Interpretation:
there is no lower-dimensional structure to uncover with PCA.
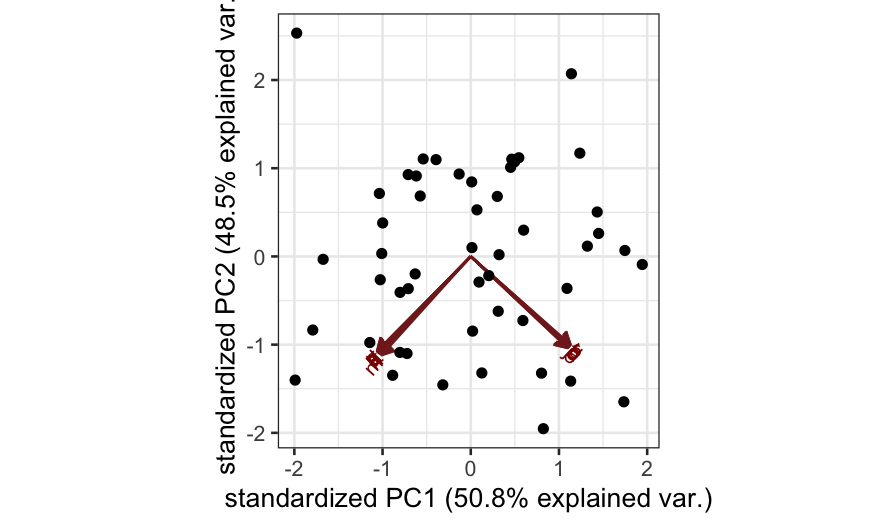
Simulation A
n = 50; p = 20
cov.mat = matrix(.99, nrow = p, ncol = p)
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = FALSE)
ggbiplot(out_prcomp)
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
Simulation B
cov.mat = matrix(0, nrow = p, ncol = p)
cov.mat[1:(p/2), 1:(p/2)] = cov.mat[(p/2 + 1):p, (p/2 + 1):p] = .99
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
Simulation C
X = matrix(rnorm(n * p), nrow = n)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
Comparing to a null distribution
- Assuming the elements of the data matrix are iid Gaussians or the
sample covariance matrix has a Wishart distribution, the limiting
distributions of the eigenvalues have been worked out.
- The empirical distribution of the eigenvalues will converge to the
Marcenko-Pastur
distribution.
- These limits are a bit different than what you see in your other
statistical theory courses, they are based on taking both the number of
samples and the number of variables to infinity in a fixed ratio.
- The package
RMTstat contains the relevant
distributions, we will use pWishartMax for the distribution
of the maximum eigenvalue of a Wishart matrix.
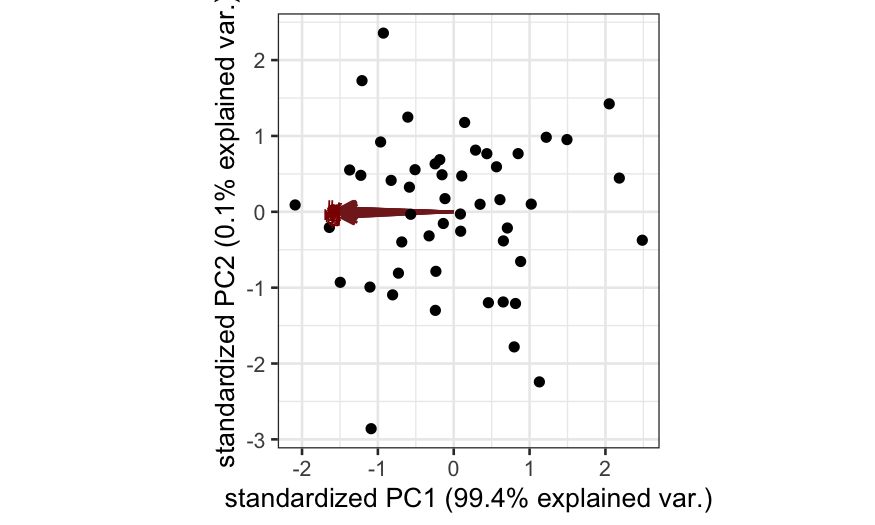
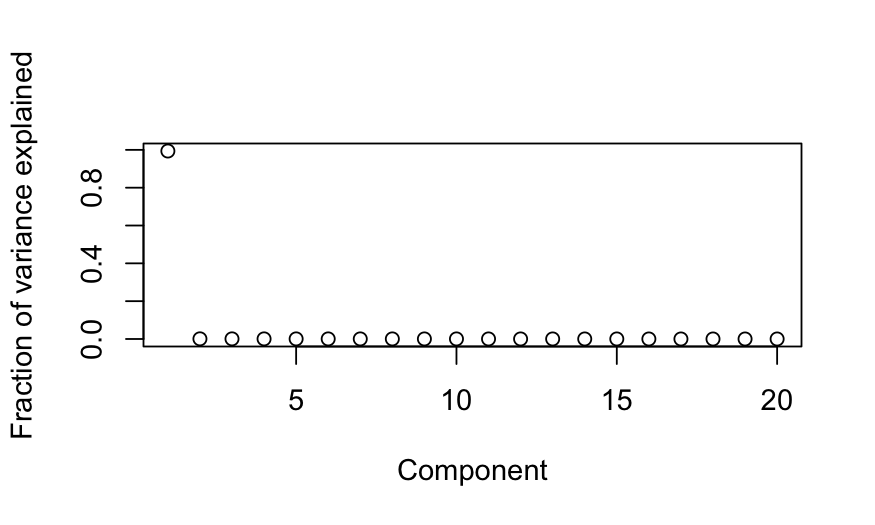
Simulation A
n = 50; p = 20
cov.mat = matrix(.99, nrow = p, ncol = p)
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = FALSE)
ggbiplot(out_prcomp)
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
pWishartMax(out_prcomp$sdev[1]^2, ndf = n, pdim = p, lower.tail = FALSE)
## [1] 0
Simulation B
cov.mat = matrix(0, nrow = p, ncol = p)
cov.mat[1:(p/2), 1:(p/2)] = cov.mat[(p/2 + 1):p, (p/2 + 1):p] = .99
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
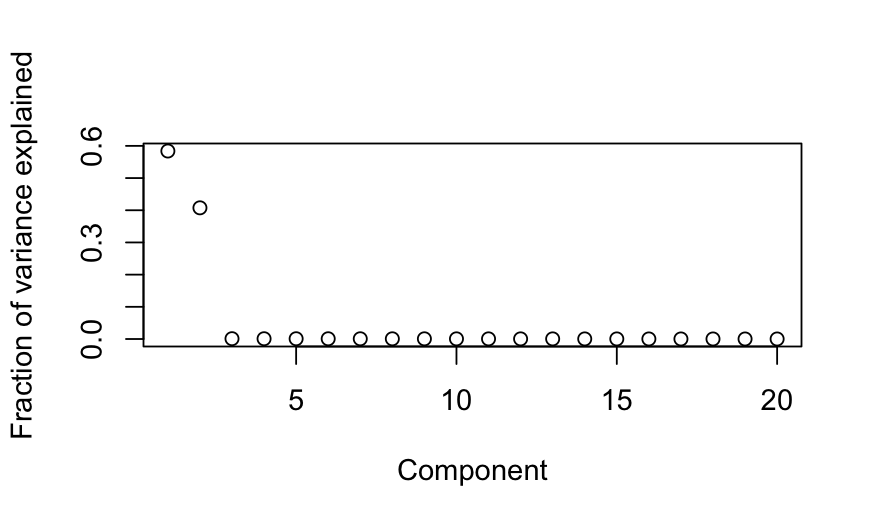
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
pWishartMax(out_prcomp$sdev[1]^2, ndf = n, pdim = p, lower.tail = FALSE)
## [1] 0
Simulation C
X = matrix(rnorm(n * p), nrow = n)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
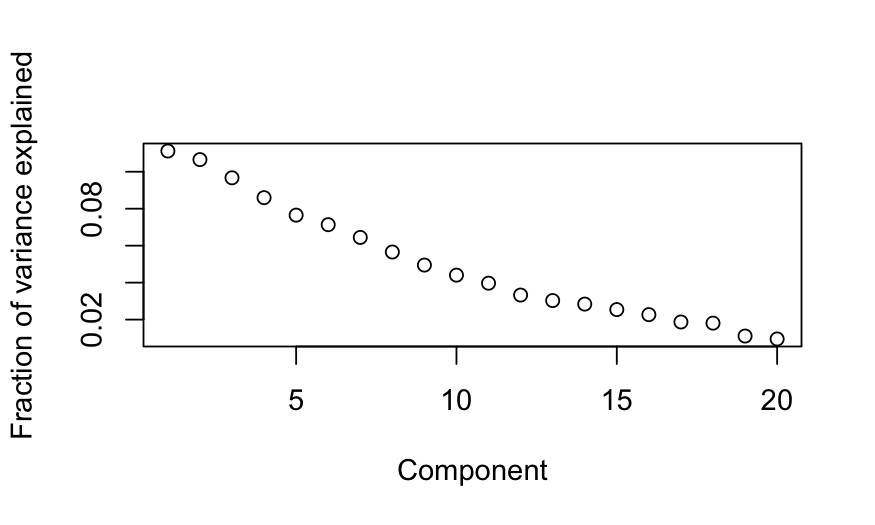
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
pWishartMax(out_prcomp$sdev[1]^2, ndf = n, pdim = p, lower.tail = FALSE)
## [1] 0.8334145
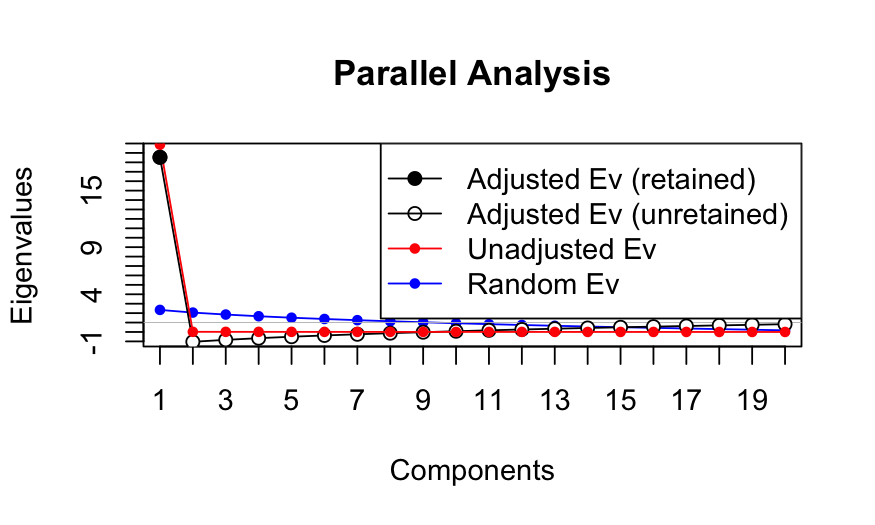
Comparing to a null distirbution: Parallel analysis
Suppose you don’t want to assume that the elements of the data matrix
are iid Gaussian, or you don’t have a large number of samples/variables.
You can use parallel analysis.
Procedure:
- Permute each of the columns of the matrix.
- Compute the eigenvalues for the permuted data matrix.
- Compare observed eigenvalues to the eigenvalues computed from the
permuted data matrix.
This procedure gives the distribution of the eigenvalues assuming
that the variables are uncorrelated and can tell you whether the
eigenvalues you observed are bigger than you would expect if all the
variables were uncorrelated.
Note: the plots will have “adjusted” eigenvalues, for a description
of these see https://alexisdinno.com/Software/files/PA_for_PCA_vs_FA.pdf
Simulation A
n = 50; p = 20
cov.mat = matrix(.99, nrow = p, ncol = p)
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = FALSE)
ggbiplot(out_prcomp)
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 600 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
## 1 18.536246 19.877931 1.341684
## --------------------------------------------------
##
## Adjusted eigenvalues > 1 indicate dimensions to retain.
## (1 components retained)
Simulation B
cov.mat = matrix(0, nrow = p, ncol = p)
cov.mat[1:(p/2), 1:(p/2)] = cov.mat[(p/2 + 1):p, (p/2 + 1):p] = .99
diag(cov.mat) = 1
X = rmvnorm(n = n, sigma = cov.mat)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
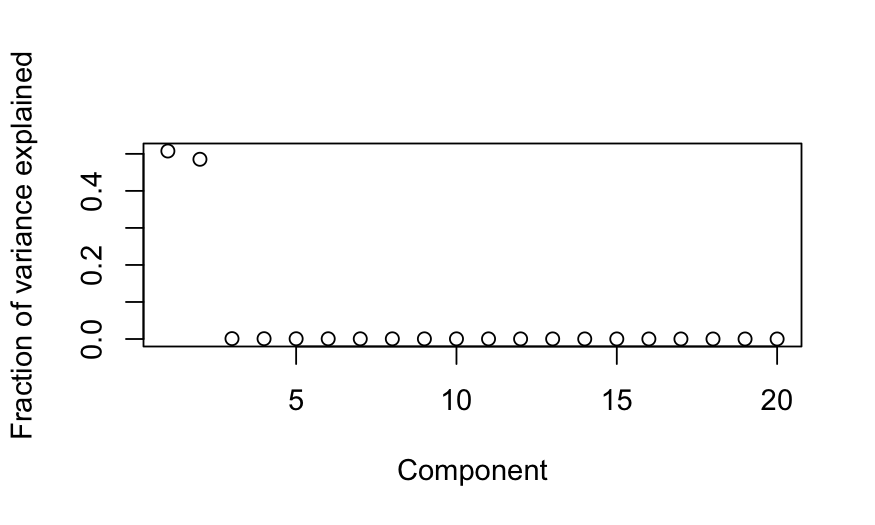
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 600 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
## 1 8.801628 10.153689 1.352060
## 2 8.654407 9.707843 1.053435
## --------------------------------------------------
##
## Adjusted eigenvalues > 1 indicate dimensions to retain.
## (2 components retained)
Simulation C
X = matrix(rnorm(n * p), nrow = n)
colnames(X) = letters[1:p]
out_prcomp = prcomp(X, center = TRUE, scale. = TRUE)
ggbiplot(out_prcomp)
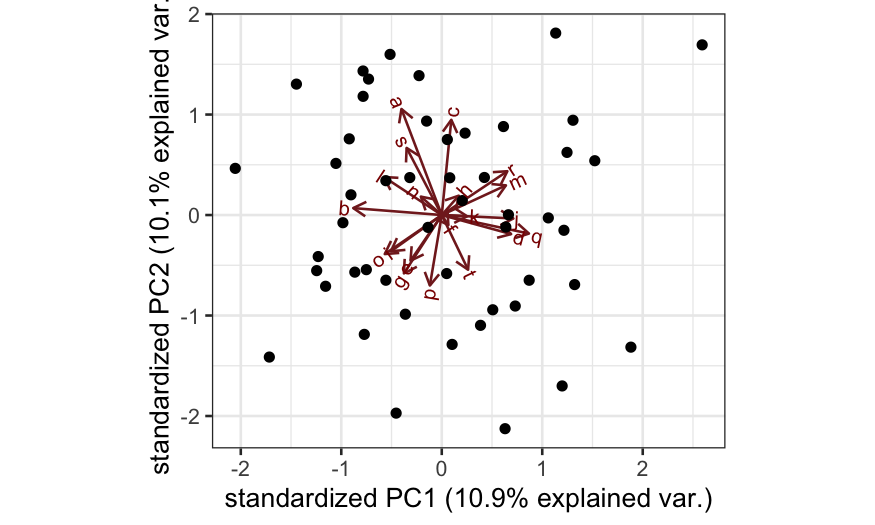
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 600 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
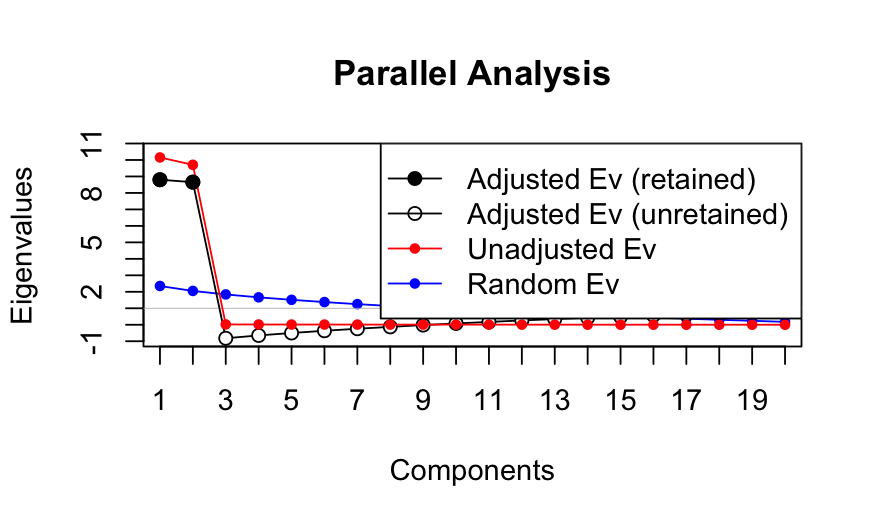
## 1 0.842456 2.181949 1.339493
## Error in if (AdjEv[x] >= 0) {: argument is of length zero
Example: Congressional voting data
A dataset containing information about votes taken by Senators in the
1999 session.
Information split across three different csvs, one giving the votes,
one giving information about the members, and one giving information
about the bills being voted on.
# read in the three data sets
vote_descriptions = read.csv("../../datasets/congress_1999/bills.csv")
members = read.csv("../../datasets/congress_1999/members.csv")
votes = read.csv("../../datasets/congress_1999/votes.csv")
## match the column names of votes and the vote_id column in vote_descriptions
vote_descriptions = vote_descriptions %>%
mutate(vote_id = str_replace(vote_id, "-", "."))
## match names and orders for members and votes
joined = join(members, votes, by = "id")
votes = joined[,(-1:-6)]
members = joined[,1:6]
## Look at the vote_descriptions data frame
table(vote_descriptions$category)
##
## amendment cloture conviction nomination passage procedural treaty
## 103 36 2 10 61 161 1
We would like to perform PCA, so we need te recode the votes from
Yea/Nay to a number.
## Decide how to recode the variables
recode_votes = function(vote) {
if(is.na(vote)) {
return(0)
} else if(vote == "Yea") {
return(1)
} else if(vote == "Nay") {
return(-1)
} else {
return(0)
}
}
votes_numeric = apply(votes, 1:2, recode_votes)
## These votes are going to cause problems for parallel analysis, so we remove them
votes_to_remove = which(apply(votes_numeric, 2, sd) == 0)
votes_numeric = votes_numeric[,-votes_to_remove]
vote_descriptions = vote_descriptions[-votes_to_remove,]
## PCA
out_prcomp = prcomp(votes_numeric)
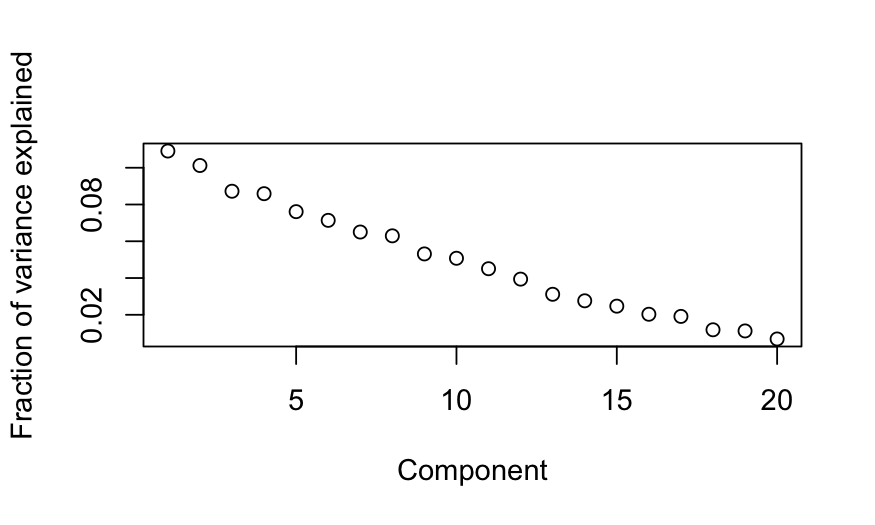
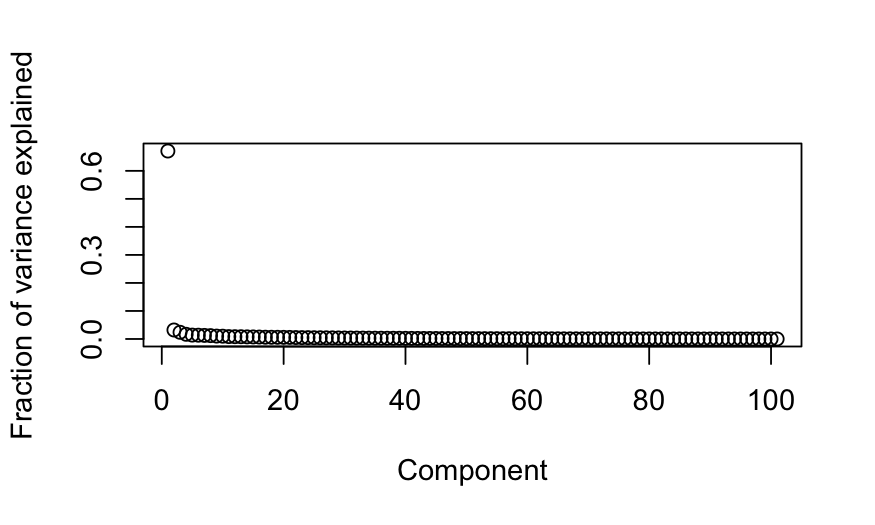
## scree plot
plot(out_prcomp$sdev^2 / sum(out_prcomp$sdev^2),
xlab = "Component", ylab = "Fraction of variance explained")
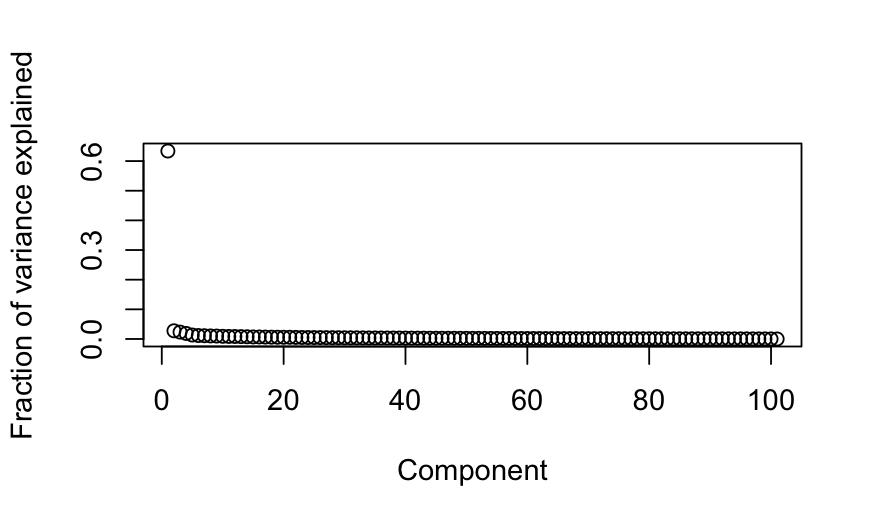
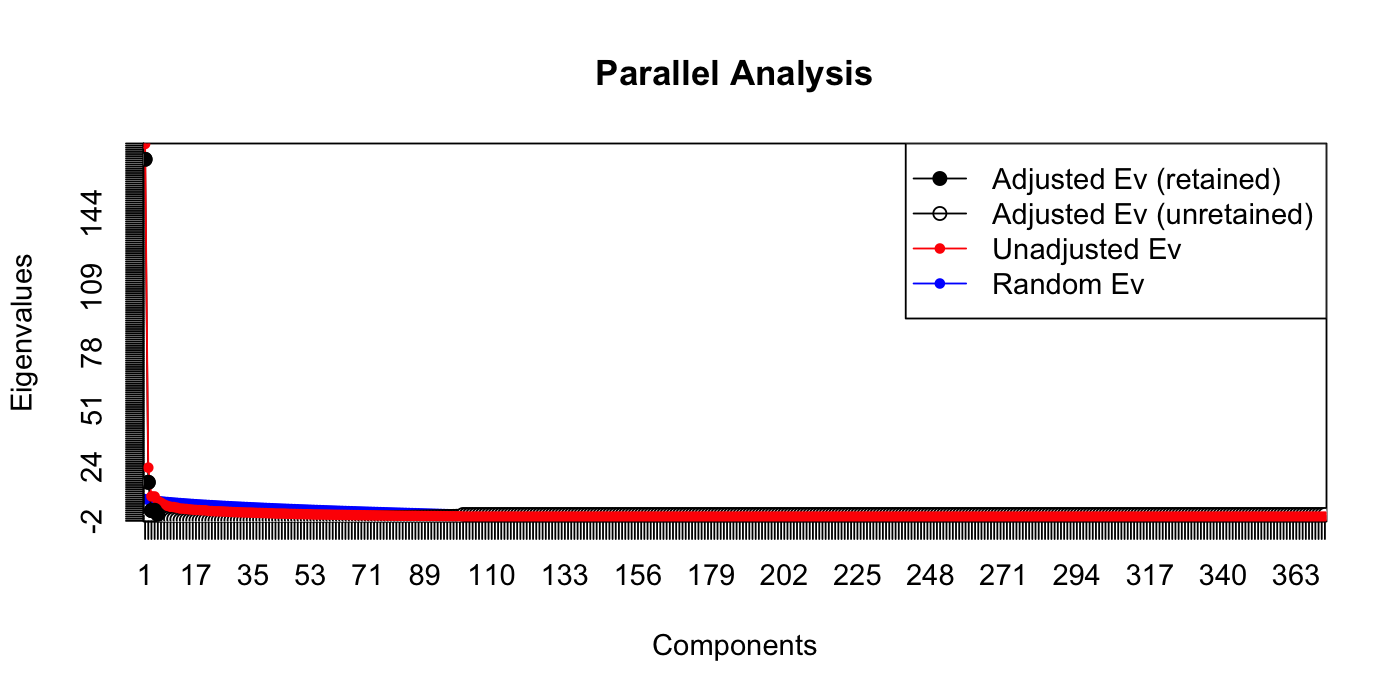
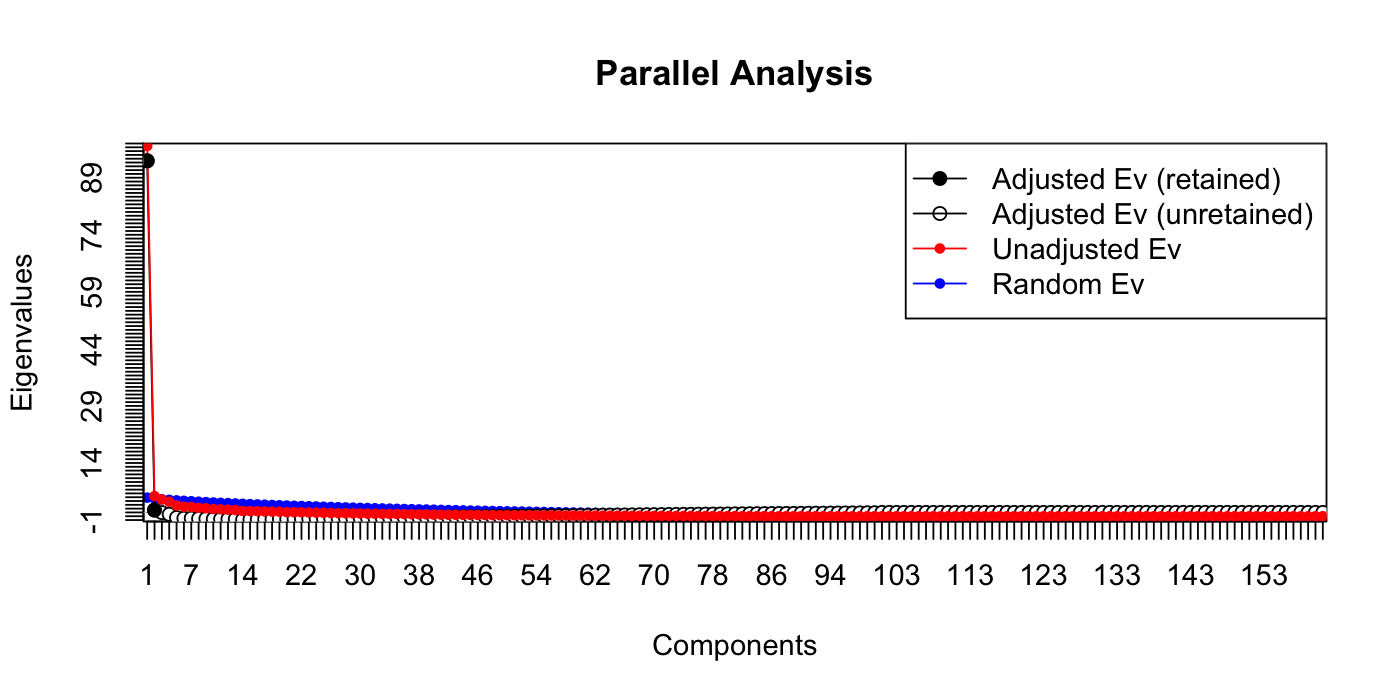
paran(votes_numeric, graph = TRUE, iterations = 100)
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 100 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
## 1 169.50319 176.781706 7.278510
## 2 16.274826 23.231897 6.957071
## 3 2.875923 9.615791 6.739868
## 4 3.023778 9.557597 6.533818
## 5 1.109663 7.472130 6.362466
## --------------------------------------------------
##
## Adjusted eigenvalues > 1 indicate dimensions to retain.
## (5 components retained)
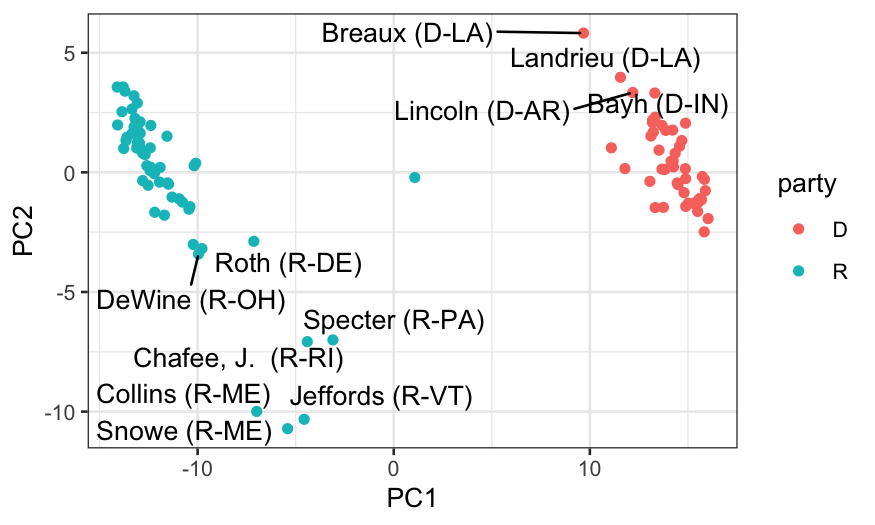
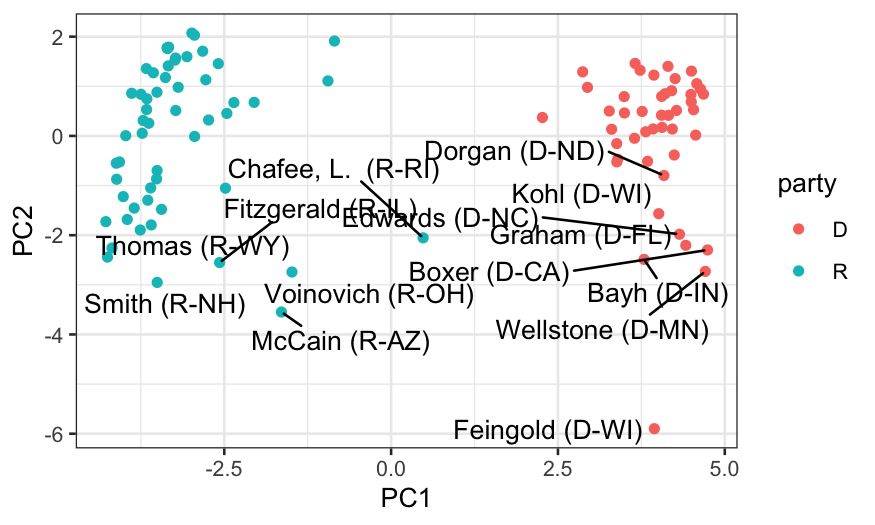
## let's look at who the senators are
members_and_scores = data.frame(members, out_prcomp$x)
ggplot(members_and_scores, aes(x = PC1, y = PC2)) +
geom_point(aes(color = party)) +
geom_text_repel(aes(label = display_name))
Looking at just the “passage” votes
out_prcomp_passage = prcomp(votes_numeric[,vote_descriptions$category == "passage"])
plot(out_prcomp_passage$sdev^2 / sum(out_prcomp_passage$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
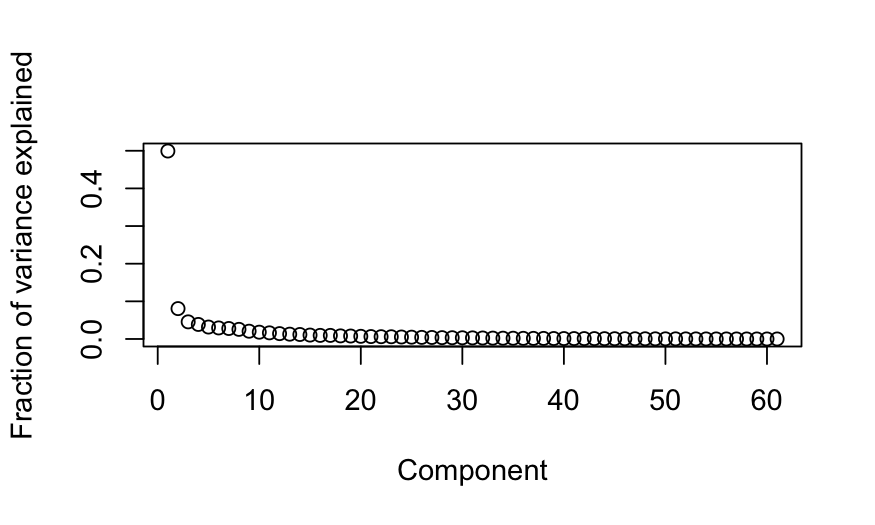
out_prcomp_passage$sdev[1]^2 / sum(out_prcomp_passage$sdev^2)
## [1] 0.499316
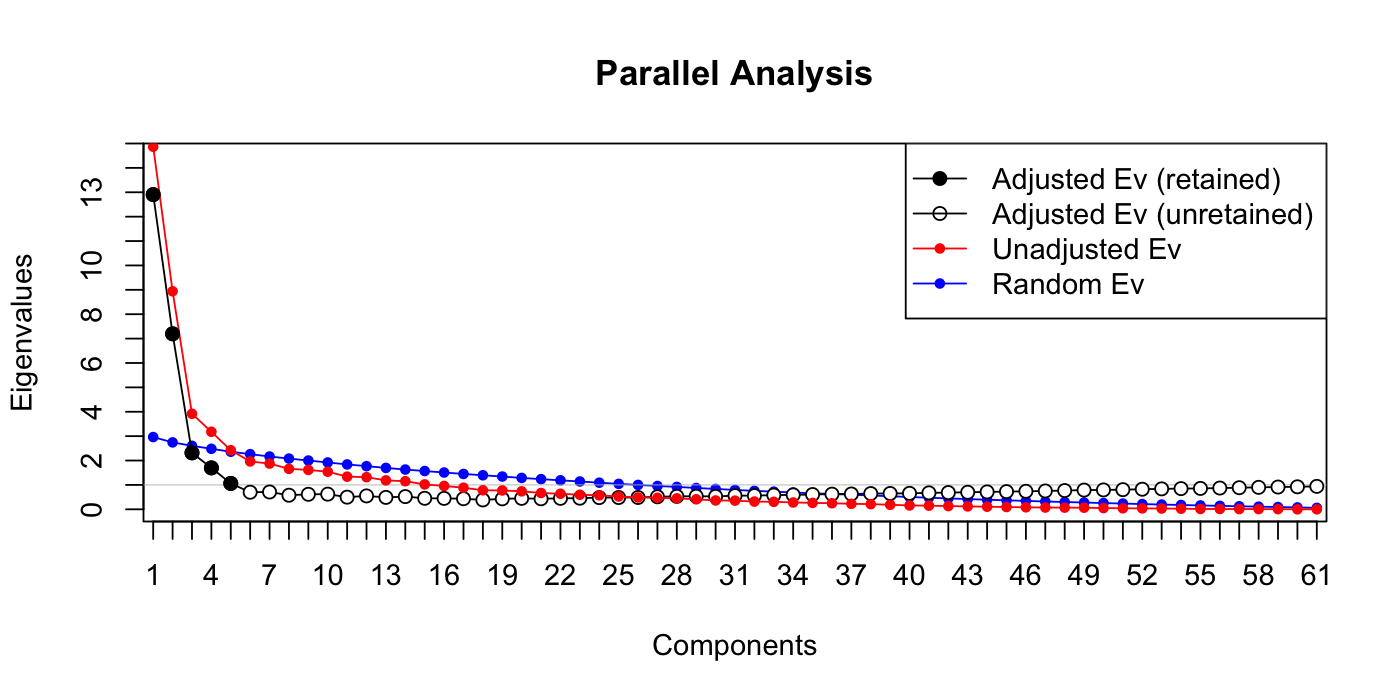
paran(votes_numeric[,vote_descriptions$category == "passage"], graph = TRUE, iterations = 100)
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 100 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
## 1 12.904483 14.866993 1.962509
## 2 7.196930 8.941067 1.744137
## 3 2.314958 3.918645 1.603687
## 4 1.698357 3.179481 1.481123
## 5 1.061265 2.422535 1.361270
## --------------------------------------------------
##
## Adjusted eigenvalues > 1 indicate dimensions to retain.
## (5 components retained)
members_and_scores_passage = data.frame(members, out_prcomp_passage$x)
ggplot(members_and_scores_passage, aes(x = PC1, y = PC2)) +
geom_point(aes(color = party)) +
geom_text_repel(aes(label = display_name))
Looking at just the “procedural” votes
out_prcomp_procedural = prcomp(votes_numeric[,vote_descriptions$category == "procedural"])
plot(out_prcomp_procedural$sdev^2 / sum(out_prcomp_procedural$sdev^2),
ylab = "Fraction of variance explained", xlab = "Component")
out_prcomp_procedural$sdev[1]^2 / sum(out_prcomp_procedural$sdev^2)
## [1] 0.670782
paran(votes_numeric[,vote_descriptions$category == "procedural"], graph = TRUE, iterations = 100)
##
## Using eigendecomposition of correlation matrix.
## Computing: 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
##
##
## Results of Horn's Parallel Analysis for component retention
## 100 iterations, using the mean estimate
##
## --------------------------------------------------
## Component Adjusted Unadjusted Estimated
## Eigenvalue Eigenvalue Bias
## --------------------------------------------------
## 1 93.440562 97.330067 3.889504
## 2 1.622993 5.285660 3.662667
## --------------------------------------------------
##
## Adjusted eigenvalues > 1 indicate dimensions to retain.
## (2 components retained)
members_and_scores_procedural = data.frame(members, out_prcomp_procedural$x)
ggplot(members_and_scores_procedural, aes(x = PC1, y = PC2)) +
geom_point(aes(color = party)) +
geom_text_repel(aes(label = display_name))
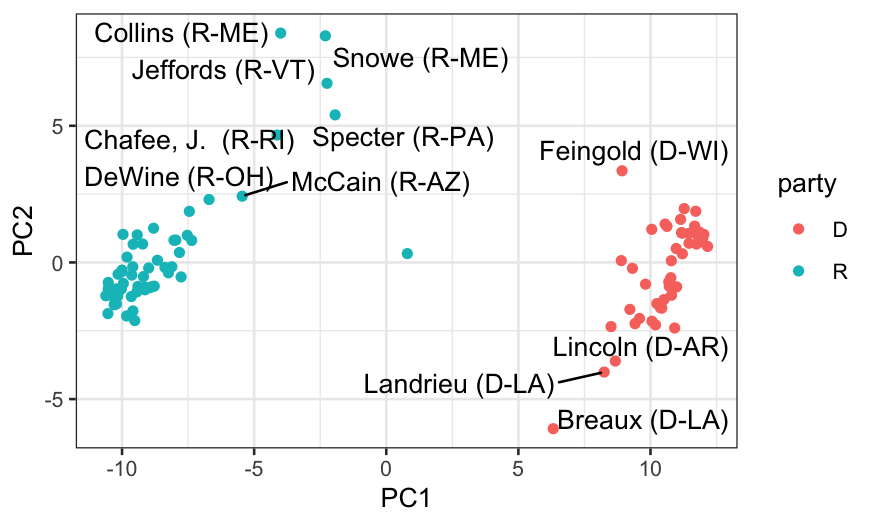
What did we learn?
- We always see one dimension that explains a very large fraction of
the variance.
- In both the votes overall and in the passage votes, we get five
dimensions that have more information than we would expect if all the
votes were uncorrelated with each other.
- In just the procedural votes, the first dimension explains more of
the variance and there are only two dimensions that have more
information than we would expect if all the votes were uncorrelated with
each other.
Overall
- Important to look at the scree plot
- Comparing to a null distribution can help with interpretation.
Parallel analysis is probably the most useful, but you can also use the
random matrix theory-based results as well.
- Parallel analysis usually more accurate.




























