Bayesian Statistics
Suppose we have data \(y_1,\ldots,
y_n\) that we believe can be described by a probability model
with parameters \(\theta\).
We also have a prior distribution on the parameters \(\theta\), describing our belief about the
values of those parameters before seeing any of the data.
\[
\begin{align*}
y_i \mid \theta &\sim P(y_i \mid \theta), \quad i = 1,\ldots, n\\
\theta & \sim \pi
\end{align*}
\]
Posterior distribution
By applying Bayes’ rule, we can compute the posterior
distribution of the parameters given the data: \[
\begin{align*}
P(\theta \mid y_1,\ldots, y_n) &= \frac{P(y_1,\ldots, y_n \mid
\theta)P(\theta)}{P(y_1,\ldots, y_n)}
\end{align*}
\]
- We want to know as much about this distribution as possible.
- For simple cases it is available in closed form
- For more complicated cases our best hope is to draw samples of
it
- From those samples we can estimate posterior means, variances,
etc.
One way of drawing samples from the posterior
Inputs:
- A target posterior: \(P(\theta \mid
y_{\text{obs}}) \propto P(y_{\text{obs}} \mid \theta)
P(\theta)\)
- A way of simulating from \(P(y_{\text{obs}} \mid \theta)\)
- A prior on the parameters \(P(\theta)\) and a way of simulating from
\(P(\theta)\)
Sampling: for \(i = 1,\ldots,
N\):
- Generate \(\theta^{(i)} \sim
P(\theta)\)
- Generate \(y^{(i)} \sim P(y \mid
\theta^{(i)})\)
- If \(y^{(i)} = y_{\text{obs}}\),
accept \(\theta^{(i)}\)
Why does this work?
- Our draws \((\theta^{(i)},
y^{(i)})\) are samples from the joint distribution \(P(\theta, y)\)
- Keeping only the values for which \(y^{(i)} = y_{\text{obs}}\) is the
definition of conditioning on \(y_{\text{obs}}\).
(Non-)Approximate Bayesian Computation Simple Example
Bayesian analysis for a Gamma-Poisson model:
Model is: \[
\begin{align*}
Y_i &\sim \text{Poisson}(\theta), \quad i = 1,\ldots, n \\
\theta &\sim \text{Gamma}(\alpha, \beta)
\end{align*}
\]
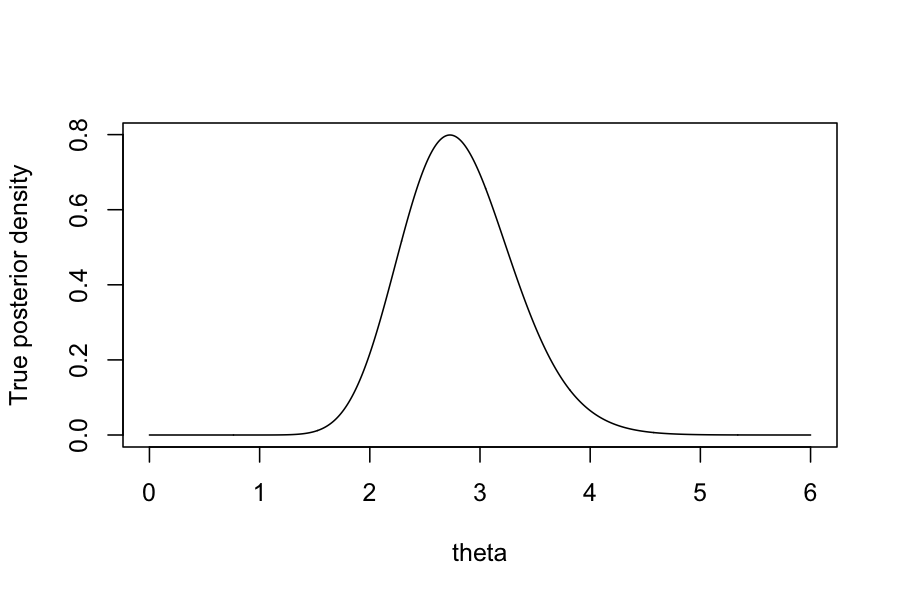
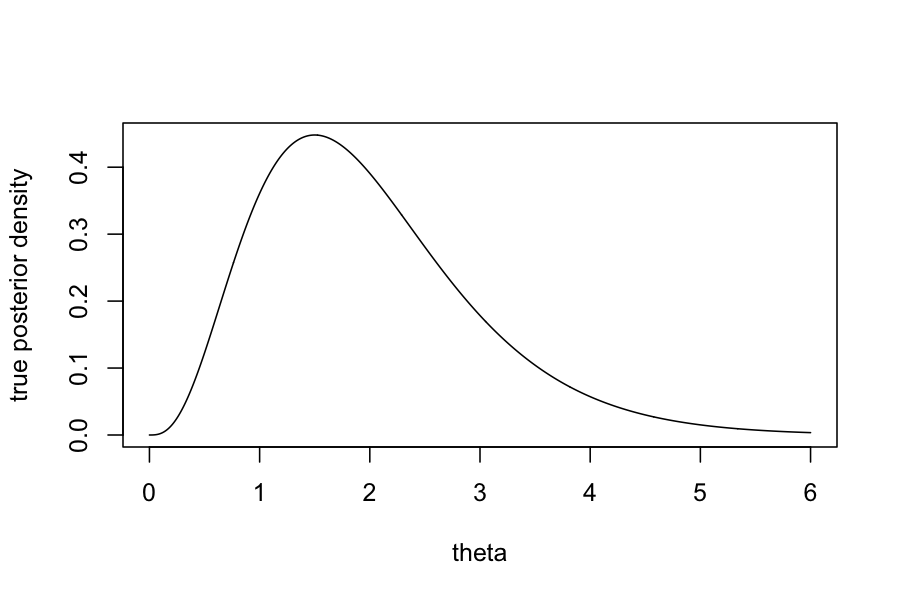
By Bayes rule, we can find in closed form that the posterior, \(P(\theta \mid Y_1, \ldots, Y_n)\) has a
\(\text{Gamma}(\sum_{i=1}^n Y_i + \alpha, n +
\beta)\) distribution.
Let’s pretend we can’t do that though, and try using the sampling
method we described on the previous slide.
Set up a function for the approximate version of ABC:
generate_abc_sample <- function(observed_data,
summary_statistic,
prior_distribution,
data_generating_function,
epsilon) {
while(TRUE) {
theta <- prior_distribution()
y <- data_generating_function(theta)
if(abs(summary_statistic(y) - summary_statistic(observed_data)) < epsilon) {
return(theta)
}
}
}
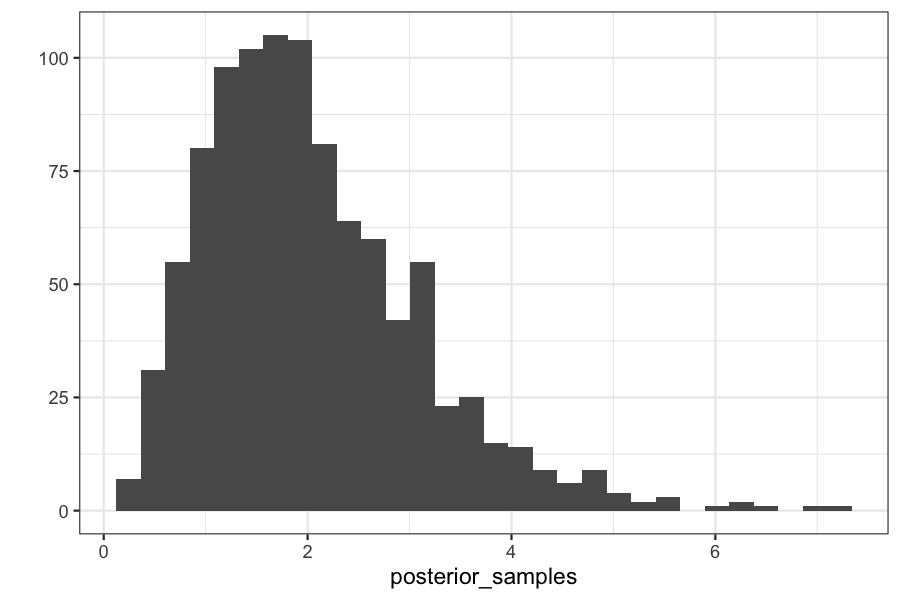
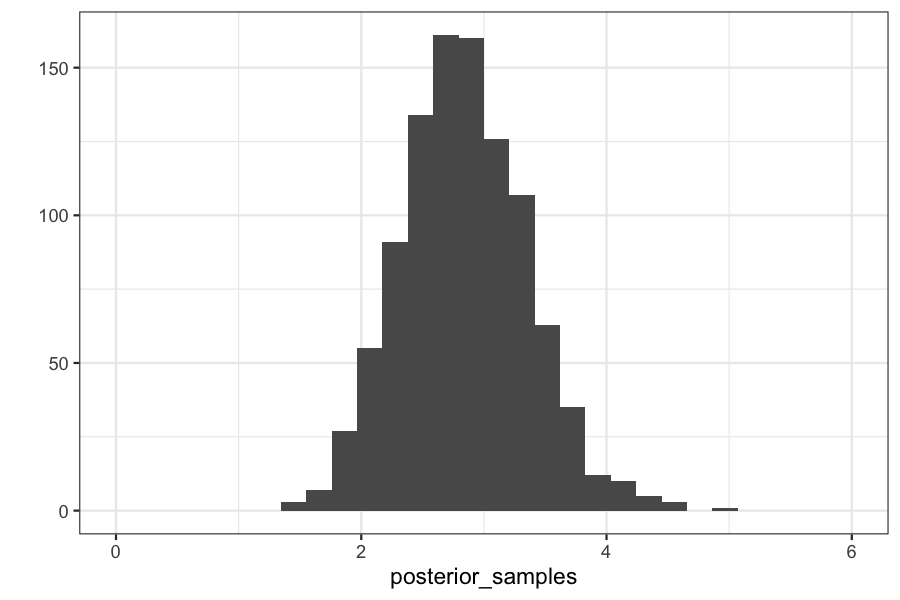
Checking on the posterior distributions:
qplot(posterior_samples) + xlim(c(0, max(theta_vec)))
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
plot(dgamma(theta_vec, shape = 1 + sum(observed_data), rate = n_samples + 1) ~ theta_vec, type = 'l', ylab = "True posterior density", xlab = "theta")