Variations on Newton’s Method
Agenda today
Newton’s method for multivariate problems
Generalized linear models and exponential familiies
Iteratively reweighted least squares for maximum likelihood in
generalized linear models
From last time
Newton’s method:
Pick some starting value \(\theta^{(0)}\)
Iterate: \(\theta^{(m+1)} =
\theta^{(m)} - d^2 \ell(\theta^{(m)}) d
\ell(\theta^{(m)})\)
Stop when \(d\ell(\theta^{(m)})\) is sufficiently close
to 0.
Newton’s method for linear regression
Linear regression model:
- \(y \in \mathbb R^n\),
- \(X \in \mathbb R^{n \times
p}\),
- \(\theta \in \mathbb R^p\),
- \(\sigma \in \mathbb R^+\)
- \(y \sim N(X \theta,
\sigma^2)\)
Likelihood: \[
L(\theta) = (2\pi)^{-n/2} \sigma^{-n}\exp(-(y - X\theta)^T (y - X\theta)
/ 2)
\]
Log likelihood: \[
\ell(\theta) = -n \log(2\pi) / 2 - n \log \sigma - (y - X \theta)^T (y -
X \theta) / 2
\]
\(d\ell(\theta)\): \[
X^T (y - X \theta)
\]
\(d^2 \ell(\theta)\): \[
-X^T X
\]
Newton step: \[
\begin{align*}
\theta^{(1)} &= \theta^{(0)} - d^2 \ell(\theta^{(0)})^{-1} d
\ell(\theta^{(0)})\\
&= \theta^{(0)} - (-X^T X)^{-1} X^T(y - X \theta^{(0)}) \\
&= \theta^{(0)} + (X^T X)^{-1} X^T y - (X^T X)^{-1} X^T X
\theta^{(0)} \\
&= (X^T X)^{-1} X^T y
\end{align*}
\]
Newton’s method for logistic regression
Logistic regression model:
- \(y \in \{0,1\}^n\),
- \(x_i \in \mathbb R^p\), \(X \in \mathbb R^{n \times p}\), \(X = \begin{pmatrix} x_1^T \\ \vdots \\ x_n^T
\end{pmatrix}\)
- \(\theta \in \mathbb R^p\),
- \(y_i \sim \text{Bern}(p_i)\),
\(p_i=\exp(x_i^T \theta) / (1 + \exp(x_i^T
\theta))\)
Likelihood: \[
L(\theta) = \prod_{i=1}^n p_i^{y_i} (1 - p_i)^{1 - y_i}
\]
Log likelihood: \[
\begin{align*}
\ell(\theta)&= \sum_{i=1}^n (y_i \log p_i + (1 - y_i) \log(1 - p_i))
\\
&= \sum_{i=1}^n (y_i x_i^T \theta - \log(1 + \exp(x_i^T \theta)))
\end{align*}
\]
First derivatives: \[
\begin{align*}
d\ell(\theta) &= \sum_{i=1}^n \left(y_i x_i - \frac{\exp(x_i^T
\theta)}{1 + \exp(x_i^T \theta)} x_i\right) \\
&= \sum_{i=1}^n (y_i - p_i) x_i \\
&= X^T (y - p)
\end{align*}
\]
Second derivatives: \[
\begin{align*}
d^2 \ell(\theta) &= -\sum_{i=1}^n p_i(1 - p_i) x_i x_i^T \\
&= -X^T W X
\end{align*}
\] if \(W = \text{diag}(p_1(1 - p_1),
\ldots, p_n(1 - p_n))\)
Newton step: \[
\begin{align*}
\theta^{(m+1)} &= \theta^{(m)} - (d^2 \ell(\theta))^{-1} d
\ell(\theta) \\
&= \theta^{(m)} + (X^T W^{(m)} X)^{-1} X^T (y - p^{(m)})
\end{align*}
\]
Translate the previous two slides into R code:
log_likelihood = function(theta, X, y) {
p = exp(X %*% theta)/ (1 + exp(X %*% theta))
return(sum(y * log(p) + (1 - y) * log(1 - p)))
}
d_log_likelihood = function(theta, X, y) {
p = exp(X %*% theta) / (1 + exp(X %*% theta))
return(t(X) %*% (y - p))
}
d2_log_likelihood = function(theta, X, y) {
p = exp(X %*% theta) / (1 + exp(X %*% theta))
W = diag(as.vector(p * (1 - p)))
return(-t(X) %*% W %*% X)
}
newton_update = function(theta_start, d_log_likelihood, d2_log_likelihood, X, y) {
step = solve(d2_log_likelihood(theta_start, X, y)) %*% d_log_likelihood(theta_start, X, y)
return(theta_start - step)
}
Make some example data
set.seed(1)
theta_true = c(1,2)
n = 100
X = cbind(rep(1, n), rnorm(n, mean = 0, sd = 4))
colnames(X) = c("Intercept", "x")
p = exp(X %*% theta_true) / (1 + exp(X %*% theta_true))
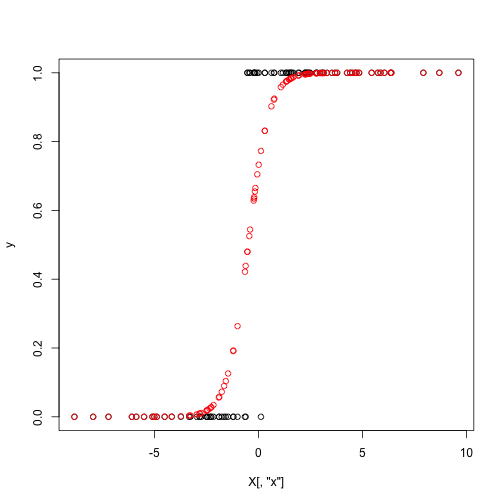
y = rbinom(n = n, size = 1, prob = p)
plot(y ~ X[,"x"])
points(p ~ X[,"x"], col = "red")
Perform Newton’s method for 10 iterations:
n_iter = 10
theta_start = c(0,0)
theta = theta_start
for(i in 1:n_iter) {
theta = newton_update(theta, d_log_likelihood, d2_log_likelihood, X, y)
cat(sprintf("Value of theta at iteration %i: (%.2f, %.2f)\n", i, theta[1], theta[2]))
}
## Value of theta at iteration 1: (0.34, 0.42)
## Value of theta at iteration 2: (0.58, 0.81)
## Value of theta at iteration 3: (0.91, 1.31)
## Value of theta at iteration 4: (1.35, 1.95)
## Value of theta at iteration 5: (1.88, 2.76)
## Value of theta at iteration 6: (2.39, 3.66)
## Value of theta at iteration 7: (2.76, 4.40)
## Value of theta at iteration 8: (2.91, 4.73)
## Value of theta at iteration 9: (2.93, 4.78)
## Value of theta at iteration 10: (2.93, 4.78)
We can check against what the glm function tells us:
glm(y ~ 0 + X, family = "binomial")
## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
##
## Call: glm(formula = y ~ 0 + X, family = "binomial")
##
## Coefficients:
## XIntercept Xx
## 2.934 4.779
##
## Degrees of Freedom: 100 Total (i.e. Null); 98 Residual
## Null Deviance: 138.6
## Residual Deviance: 15.26 AIC: 19.26
Look in a little more detail. Define a function that gives us \(p\) and \(W\) at an iteration:
get_fitted_and_weights = function(theta, X, y) {
p = exp(X %*% theta) / (1 + exp(X %*% theta))
W = p * (1 - p)
return(list(fitted = p, weights = W))
}
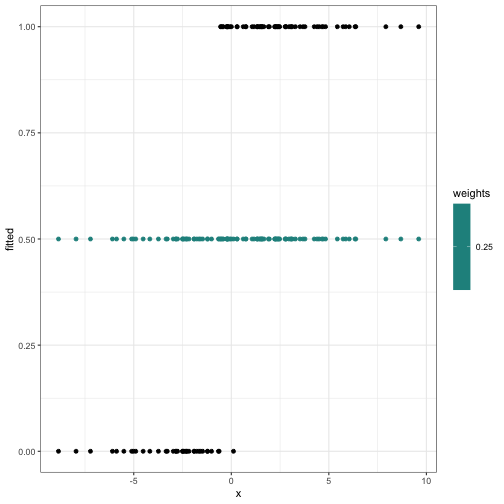
What \(W\), \(p\), \(y -
p\) look like starting out:
theta_start = c(0,0)
theta = theta_start
fitted_and_weights = get_fitted_and_weights(theta, X, y)
internals_df = data.frame(x = X[,2], y = y, fitted = fitted_and_weights$fitted, weights = fitted_and_weights$weights)
ggplot(internals_df) +
geom_point(aes(x = x, y = fitted, col = weights)) +
geom_point(aes(x = x, y = y)) +
scale_color_viridis()
ggplot(internals_df) +
geom_point(aes(x = x, y = (y - fitted) / weights, col = weights)) + scale_color_viridis()
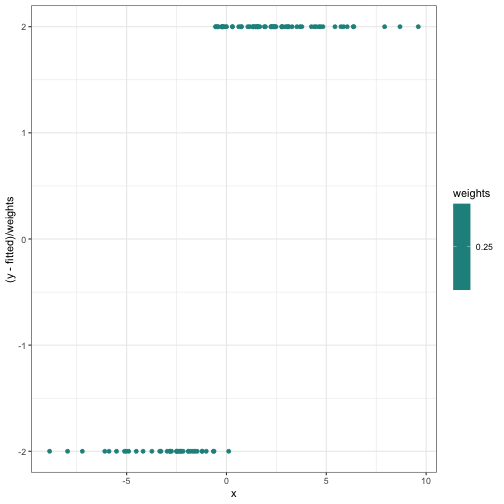
Something suspicious:
lm((y - fitted) / weights ~ x, data = internals_df, weights = weights)
##
## Call:
## lm(formula = (y - fitted)/weights ~ x, data = internals_df, weights = weights)
##
## Coefficients:
## (Intercept) x
## 0.3366 0.4212
-solve(d2_log_likelihood(theta, X, y)) %*% d_log_likelihood(theta, X, y)
## [,1]
## Intercept 0.3365656
## x 0.4211563
theta = newton_update(theta, d_log_likelihood, d2_log_likelihood, X, y)
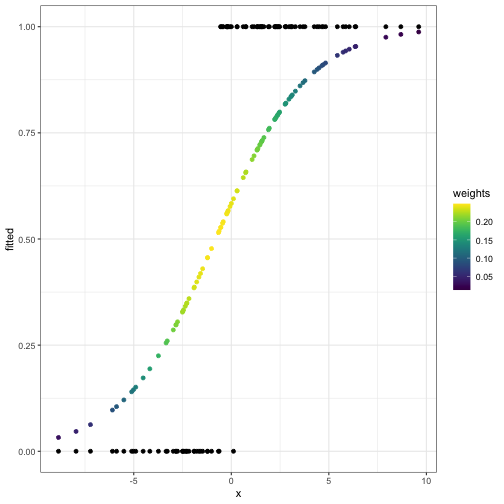
What \(W\), \(p\), \(y -
p\) look like after the first iteration:
fitted_and_weights = get_fitted_and_weights(theta, X, y)
internals_df = data.frame(x = X[,2], y = y, fitted = fitted_and_weights$fitted, weights = fitted_and_weights$weights)
ggplot(internals_df) +
geom_point(aes(x = x, y = fitted, col = weights)) +
geom_point(aes(x = x, y = y)) +
scale_color_viridis()
ggplot(internals_df) +
geom_point(aes(x = x, y = (y - fitted) / weights, col = weights)) + scale_color_viridis()
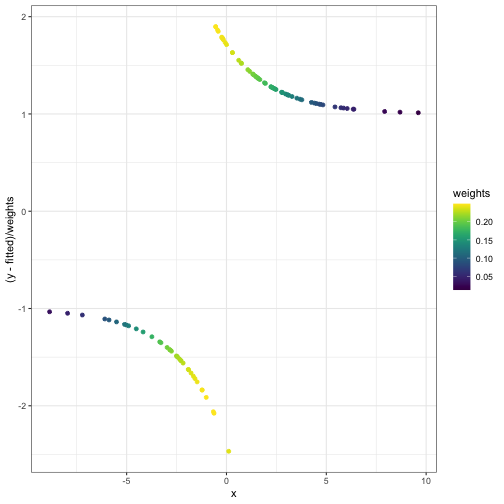
Something suspicious:
lm((y - fitted) / weights ~ x, data = internals_df, weights = weights)
##
## Call:
## lm(formula = (y - fitted)/weights ~ x, data = internals_df, weights = weights)
##
## Coefficients:
## (Intercept) x
## 0.2465 0.3863
-solve(d2_log_likelihood(theta, X, y)) %*% d_log_likelihood(theta, X, y)
## [,1]
## Intercept 0.2464510
## x 0.3863167
theta = newton_update(theta, d_log_likelihood, d2_log_likelihood, X, y)
Exponential families and generalized linear models
Exponential
families are families of probability distributions whose densities
take the form \[
f(x | \eta) = h(x) exp(\eta^T T(x) - A(\eta))
\]
\(T\) and \(A\) are known functions that describe the
family.
\(\eta\) is the natural
parameter.
Properties that we’ll need later:
Example: Bernoulli distribution
“Normal” parameterization: \(p(y \mid
p) = p^y (1 - p)^{1 - y}\)
Rewrite as \[
p(y \mid p) = \exp\left[ \log(p / (1 - p)) y + \log(1 - p) \right]
\]
Then: \[
\begin{align*}
\eta &= p / (1 - p) \\
T(y) &= y \\
A(\eta) &= -\log(1 - p) = \log(1 + e^\eta)\\
h(y) &= 1\\
\end{align*}
\]
Example: Normal distribution with just a mean
“Normal” parameterization: \(p(y \mid
\mu) = (2 \pi)^{-1/2} \exp \left[ -\frac{1}{2} (y - \mu)^2
\right]\)
Rewrite as \[
p(y \mid \mu) = \frac{1}{\sqrt{2\pi}} \exp \left[ \mu y - \frac{1}{2}
y^2 - \frac{1}{2} \mu^2\right]
\]
Then: \[
\begin{align*}
\eta &= \mu\\
T(y) &= y\\
A(\eta) &= \frac{\mu^2}{ 2} = \frac{\eta^2}{2}\\
h(y) &= \frac{1}{\sqrt{2\pi}}\exp(-y^2 / 2)
\end{align*}
\]
Example: Normal distribution with mean and variance
“Normal” parameterization: \(p(y \mid
\mu, \sigma^2) = (2 \pi \sigma^2)^{-1/2} \exp \left[
-\frac{1}{2\sigma^2} (y - \mu)^2 \right]\)
Rewrite as \[
p(y \mid \mu, \sigma^2) = \frac{1}{\sqrt{2\pi}} \exp \left[
\frac{\mu}{\sigma^2} y - \frac{1}{2\sigma^2} y^2 - \frac{1}{2\sigma^2}
\mu^2 - \log \sigma\right]
\]
Then: \[
\begin{align*}
\eta &= \begin{pmatrix} \mu / \sigma^2 \\ -1/2\sigma^2
\end{pmatrix}\\
T(y) &= \begin{pmatrix} y \\ y^2 \end{pmatrix} \\
A(\eta) &= \frac{\mu^2}{ 2 \sigma^2} + \log \sigma =
-\frac{\eta_1^2}{4\eta_2} - \frac{1}{2} \log(-2\eta_2)\\
h(y) &= \frac{1}{\sqrt{2\pi}}
\end{align*}
\]
Why do we like them?
Many commonly-used distributions: normal, exponential, Poisson,
binomial, multinomial, etc.
Easy to analyze
They describe the stochasticity in generalized linear
models
Generalized linear models
Models for independent observations, \(y_i,
i = 1,\ldots, n\)
Three components:
- Random component: \[
y_i \sim f(\eta_i)
\] where \(f\) is an exponential
family, \(\eta_i\) the natural
parameter
- Systematic component or the linear predictor \[
\eta_i = x_i^T \theta
\]
- Link function: links the mean to the linear predictor \[
\eta_i = g(\mu_i)
\] \(g\) is the link function.
The relationship between the means and the predictors is then \[
\mu_i = g^{-1} (x_i^T \theta)
\]
Choice of link functions
The canonical link is the one that maps the mean to the
natural parameter. If you use the canonical link, the natural parameter
will be linear in the predictors.
Normal: Canonical link is the identity: \(g(x) = x\)
Bernoulli: Canonical link is logit: \(g(x) = \log(x / (1 - x))\)
Poisson: Canonical link is the log: \(g(x) = \log(x)\)
Gamma: Canonical link is inverse: \(g(x) = x^{-1}\)
Iteratively Reweighted Least Squares
- Start with an estimate of the parameters \(\theta^{(0)}\).
Find \(\eta_i^{(m)} = x_i^T
\theta^{(m)}\), \(i = 1,\ldots,
n\)
Find \(\mu_i^{(m)} = g^{-1}
(\eta_i^{(m)})\)
- Compute the vector \(z^{(m)} \in \mathbb
R^n\) of “working dependent variables”: \[
z_i^{(m)} = \eta_i^{(m)} + (y_i - \mu_i^{(m)}) d\eta_i / d \mu_i
\]
- Compute iterative weights: \[
w_i^{(m)} = (A''(\eta_i^{(m)}) (d\eta_i / d \mu_i)^2)^{-1}
\] and let \(W^{(m)} \in \mathbb R^{n
\times n}\) be a diagonal matrix with \(W^{(m)}_{ii} = w_i^{(m)}\)
- Obtain \(\theta^{(m+1)}\) by
regressing the working dependent variable \(z_i\) on the predictors \(x_i\) using weights \(w_i\): \[
\theta^{(m+1)} = (X^T W^{(m)} X)^{-1} X^T W^{(m)} z^{(m)}
\]
- Iterate until convergence
Idea:
The problem is linear in the natural parameter space, so try to
do least squares there
“Working dependent variable” is like \(y_i\) mapped to the natural parameter
space.
In general, a random variable taken from an exponential family
distribution will have variance that depends on the natural
parameter.
The weights are inversely proportional to the variance of the
working dependent variable at the current guess for \(\theta\).
Samples for which the variance should be smaller have larger
weights, samples for which the variance should be larger get smaller
weights
Example: Linear regression
Random component: normal distribution, \(y_i \sim N(\eta_i, 1)\) (variance 1 for
ease of notation, everything goes through analogously with unknown
variance \(\sigma\))
Exponential family representation of the normal distribution has
\(A(\eta) = \eta^2 / 2\)
Systematic component: \(\eta_i = x_i^T
\theta\)
Canonical link function for the normal distribution is \(g(x) = x\), so \(\eta_i = \mu_i\)
Identity link means \(E(y_i) =
g^{-1}(\eta_i) = g^{-1}(x_i^T \theta) = x_i^T \theta\)
- Derivative \(\frac{d\eta}{d\mu}=
1\)
Working dependent variable at iteration \(0\) \[
\begin{align*}
z_i^{(0)} &= \eta_i^{(0)} + (y_i - \mu_i^{(0)}) d \eta_i^{(0)}/
d\mu\\
&= \mu_i^{(0)} + (y _i - \mu_i^{(0)}) = y_i
\end{align*}
\]
Iterative weights: \[
\begin{align*}
w_i^{(0)} &= (A''(\eta_i^{(0)}) d\eta_i^{(0)} / d\mu)^{-1}
\\
&= 1
\end{align*}
\]
New estimate is \[
\begin{align*}
\theta^{(1)} &= (X^T W^{(0)} X)^{-1} X^T W^{(0)} z\\
&= (X^T X)^{-1} X^T y
\end{align*}
\]
Example: logistic regression
Random component: Bernoulli distribution, \(y_i \sim \text{Bernoulli}(\mu_i)\), \(\mu_i \in (0,1)\)
Systematic component: \(\eta_i = x_i^T
\theta\)
Canonical link for Bernoulli is \(g(x)
= \log(x / (1 - x))\), so \(\eta_i =
\log(\mu_i / (1 - \mu_i))\)
Exponential family representation of Bernoulli has \(A(\eta) = \log (1 + e^\eta)\)
Quantities we will need: \[
\eta = \log (\mu / (1 - \mu))
\]
\[
d\eta / d\mu = 1/\mu + 1 / (1 - \mu) = 1 / (\mu(1 - \mu))
\]
\[
A(\eta) = \log(1 + e^\eta)
\]
\[
A'(\eta) = \frac{e^\eta}{1 + e^\eta}
\]
\[
\begin{align*}
A''(\eta) &= \frac{e^\eta}{(1 + e^\eta)^2}\\
&= \mu(1 - \mu)
\end{align*}
\]
Working dependent variables: \[
z_i = \eta_i + (y_i - \mu_i) \frac{d\eta_i}{d \mu_i}\\
= \eta_i + \frac{y_i - \mu_i}{\mu_i(1 - \mu_i)}
\]
Iterative weights: \[
\begin{align*}
w_i &= (A''(\eta_i) (\frac{d\eta_i}{d\mu_i})^2)^{-1} \\
&= (\mu_i(1 - \mu_i) (\mu_i(1 - \mu_i))^{-2})^{-1}\\
&= \mu_i(1 - \mu_i)
\end{align*}
\]
Update formula:
\[
\theta^{(m+1)} = (X^T W^{(m)} X)^{-1} X^T W^{(m)} z^{(m)}
\]
Notice that we can rewrite this: \[
\begin{align*}
\theta^{(m+1)} &= (X^T W^{(m)} X)^{-1} X^T W^{(m)} z^{(m)} \\
&= (X^T W^{(m)} X)^{-1} X^T W^{(m)} (\eta^{(m)} + \frac{y^{(m)} -
\mu^{(m)}}{\mu^{(w)}(1 - \mu^{(m)})}) \\
&= (X^T W^{(m)} X)^{-1} X^T W^{(m)} (X\theta^{(m)} +
(W^{(m)})^{-1}(y - \mu^{(m)}))\\
&= \theta^{(m)} + (X^T W^{(m)} X)^{-1} X^T (y - \mu^{(m)})
\end{align*}
\]
Remember Newton-Raphson for logistic regression?
Why IRLS?
Different from Newton-Raphson if you use a non-canonical link,
e.g. probit regression instead of logistic regression.
Interpretation of Newton-Raphson suggests algorithms for other
models.
If for some reason you are in a programming environment where
weighted least squares is easy and everything else is unavailable, you
can use weighted least squares to estimate GLMs.




